SPARQLer
SPARQL
SPARQL Protocol and RDF Query Language (SPARQL) is a query language defined by the W3C standard for querying RDF data. For further information on SPARQL, see SPARQL Background.
The SPARQLer tool
The SPHN SPARQL Queries Generator
(SPARQLer) is a Python tool that accepts as input an SPHN RDF Schema
(Turtle format) and generates a series of SPARQL queries in a standard RDF/OWL
format based on the concepts present in the schema.
The tool is integrated into the SPHN Framework Schema Visualization Tool.
The automatically generated queries could be executed against a SPARQL endpoint by data managers and hospitals to retrieve the content of the RDF data in a tabular format.
Note
For more details about installation instruction and the usage of the tool, check the SPARQLer README.md.
Available SPARQL Queries
‘Concept flattening’ queries
The ‘concept flattening’ process groups all resources and their properties into one query file per concept. The data is “flattened”, in the sense that each query returns a list of resources defined for that concept together with values of the directly connected properties. If the value of a property is another concept, the values of that concept would also be retrieved. A concept flattening query can be run against a SPARQL endpoint to generate a table per concept/class.
The SPARQLer requires an RDF schema as input for the concept flattening process. The output is a SPARQL query file (.rq) for each concept, and the aim of the query is to retrieve the data as a table. This can give a better overview of the extent of metadata connected to a particular concept.
Example of concept flattening
For the Age concept, all resources of type Age
are extracted with the SubjectPseudoIdentifier object, the Quantity object
and the DeterminationDateTime value of the Age.
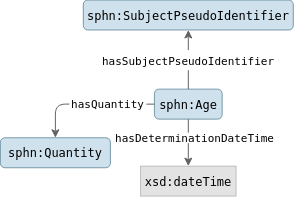
Figure 1. The concept Age and its related metadata.
The query below is the output of the SPARQLer for the Age concept:
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT *
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasIdentifier ?hasSubjectPseudoIdentifier_Identifier . }
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode ?hasSubjectPseudoIdentifier_hasDataProviderInstitute_Code . }
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasIdentifier ?hasSubjectPseudoIdentifier_hasDataProviderInstitute_hasCode_Identifier . }
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasCodingSystemAndVersion ?hasSubjectPseudoIdentifier_hasDataProviderInstitute_hasCode_CodingSystemAndVersion . }
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasName ?hasSubjectPseudoIdentifier_hasDataProviderInstitute_hasCode_Name . }
optional{ ?resource sphn:hasQuantity/sphn:hasValue ?hasQuantity_Value . }
optional{ ?resource sphn:hasQuantity/sphn:hasComparator ?hasQuantity_Comparator . }
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode ?hasQuantity_hasUnit_Code . }
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasIdentifier ?hasQuantity_hasUnit_hasCode_Identifier . }
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasCodingSystemAndVersion ?hasQuantity_hasUnit_hasCode_CodingSystemAndVersion . }
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasName ?hasQuantity_hasUnit_hasCode_Name . }
optional{ ?resource sphn:hasDeterminationDateTime ?DeterminationDateTime . }
}
The query above can be ran in any SPARQL endpoint to retrieve data about Age resources found in a database/data resource of interest that stores data in an SPHN-compliant format. The output of the query would generate a table-like representation of the concept Age and its related metadata, of which an excerpt is shown below:
resource |
hasSubjectPseudoIdentifier_Identifier |
hasQuantity_Value |
… |
|---|---|---|---|
CHE…Age-001 |
CHE…SubPseId-002 |
“20”^^xsd:double |
… |
Statistical queries
These statistical queries are manually built and based on the initially generated “hospIT statistics queries”. Prefixes, class and property names were adapted to fit the SPHN RDF Schema 2021-1. The following statistical queries are provided:
Query per concept/class: Counting the instances per concept and predicate
Query per concept/class: Minimum and maximum of predicates (dates or values)
Query per concept/class: List and count of all used codes for hasCode
etc.
Examples
The query below is the output of the SPARQLer for counting the instances of Age concept and its connected predicates:
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT *
WHERE {
{
SELECT ?origin (COUNT(?origin) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:Age" as ?origin)
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasSubjectPseudoIdentifier/sphn:hasIdentifier" as ?origin)
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasIdentifier ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode" as ?origin)
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasName" as ?origin)
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasName ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasIdentifier" as ?origin)
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasIdentifier ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasCodingSystemAndVersion" as ?origin)
optional{ ?resource sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode/sphn:hasCodingSystemAndVersion ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasDeterminationDateTime" as ?origin)
optional{ ?resource sphn:hasDeterminationDateTime ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasComparator" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasComparator ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasValue" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasValue ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasUnit/sphn:hasCode" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasName" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasName ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasIdentifier" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasIdentifier ?predicate . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (COUNT(?predicate) as ?count_instances)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasCodingSystemAndVersion" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasUnit/sphn:hasCode/sphn:hasCodingSystemAndVersion ?predicate . }
}
GROUP BY ?origin
}
}
The output of the query would generate a table-like representation of the concept Age and its predicates, of which an excerpt is shown below:
origin |
count_instances |
|---|---|
“sphn:Age” |
“10”^^xsd:integer |
“sphn:hasSubjectPseudoIdentifier/sphn:hasIdentifier” |
“10”^^xsd:integer |
“sphn:hasSubjectPseudoIdentifier/sphn:hasDataProviderInstitute/sphn:hasCode” |
“10”^^xsd:integer |
… |
… |
The query below is the output of the SPARQLer for calculating the min and max values of predicates for the Age concept:
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT *
WHERE {
{
SELECT ?origin (MIN(?value) as ?min) (MAX(?value) as ?max)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasDeterminationDateTime" as ?origin)
optional{ ?resource sphn:hasDeterminationDateTime ?value . }
}
GROUP BY ?origin
} UNION
{
SELECT ?origin (MIN(?value) as ?min) (MAX(?value) as ?max)
WHERE {
?resource a <https://biomedit.ch/rdf/sphn-ontology/sphn#Age> .
BIND("sphn:hasQuantity/sphn:hasValue" as ?origin)
optional{ ?resource sphn:hasQuantity/sphn:hasValue ?value . }
}
GROUP BY ?origin
}
}
The output of the query would generate a table-like representation of the concept Age and its hasCode predicates, of which an excerpt is shown below:
origin |
min |
max |
|---|---|---|
“sphn:hasDeterminationDateTime” |
“2022-04-06T11:55:43.673Z”^^xsd:dateTime |
“2022-05-27T11:55:43.808Z”^^xsd:dateTime |
“sphn:hasQuantity/sphn:hasValue” |
“0.03431341828097245”^^xsd:double |
“0.9737777086827694”^^xsd:double |
Availability and usage rights
© Copyright 2024, Personalized Health Informatics Group (PHI), SIB Swiss Institute of Bioinformatics.
The SPARQLer is available at (send request to DCC - dcc@sib.swiss):
Version 1.0 (supports SPHN 2021-1 and SPHN 2021-2).
Version 2.0 (supports SPHN 2021-1 and SPHN 2021-2).
Version 3.0 (supports SPHN 2021-1 to SPHN 2022-2)
Version 4.0 (supports SPHN 2021-1 to SPHN 2023-2)
The SPARQL queries for the SPHN RDF Schema 2021-2, 2022-2 and 2023-2 are available at sparql-queries.
The SPARQLer is licensed under the GPLv3 (see License).
For any question or comment, please contact the Data Coordination Center (DCC) at dcc@sib.swiss.