Querying data with SPARQL
Note
To find out more watch the Querying Data with SPARQL Training
How to setup inference in GraphDB
Here, we demonstrate how to setup inference in GraphDB. For an overview of the options to load RDF data into GraphDB, please refer to loading instructions or GraphDB’s documentation.
Creating a new repository
In order to be able to use inference capabilities of GraphDB, inference needs to be enabled when creating a new repository, as this currently can not be done afterwards. Note that a default inference is already enabled, as shown in Figure 1.
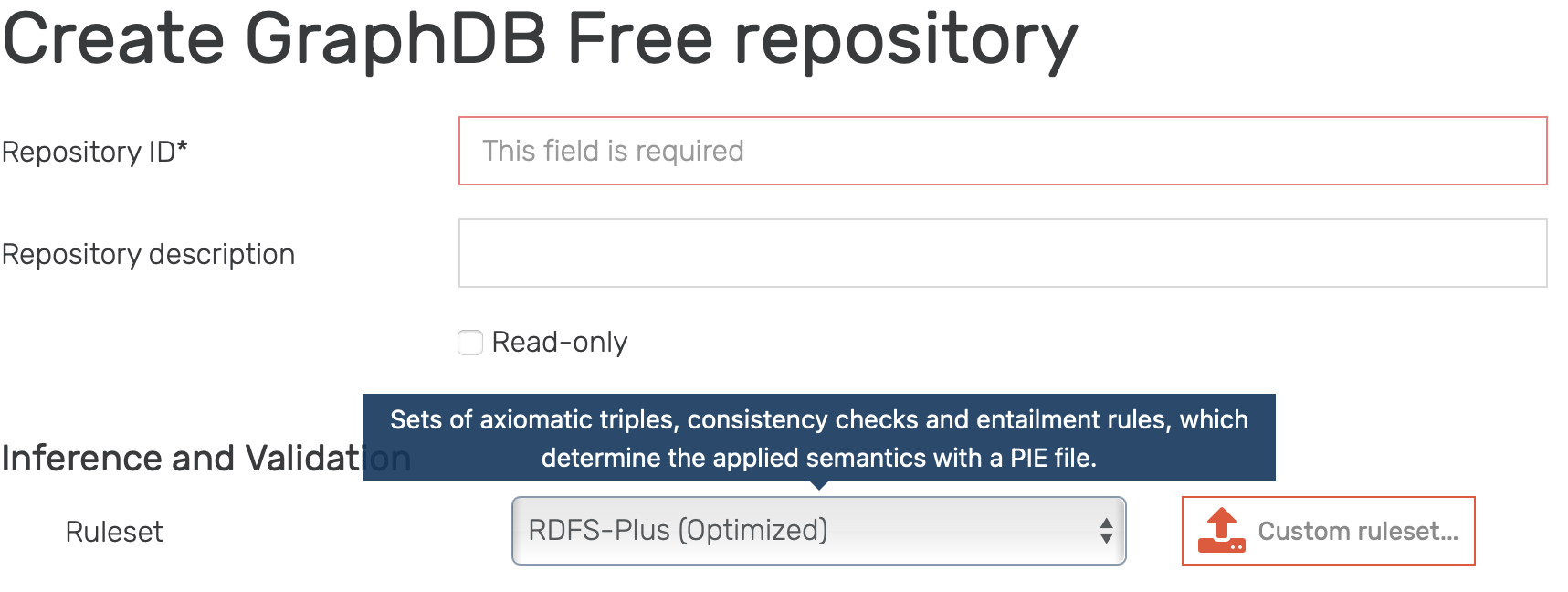
Figure 1: Creating a new repository with default inference enabled.
User settings
Following the creation of the repository with inference enabled, one can choose to exclude/include inferred data in results by selecting the corresponding option in the SPARQL editor settings (see Figure 2 and GraphDB documentation for more information).
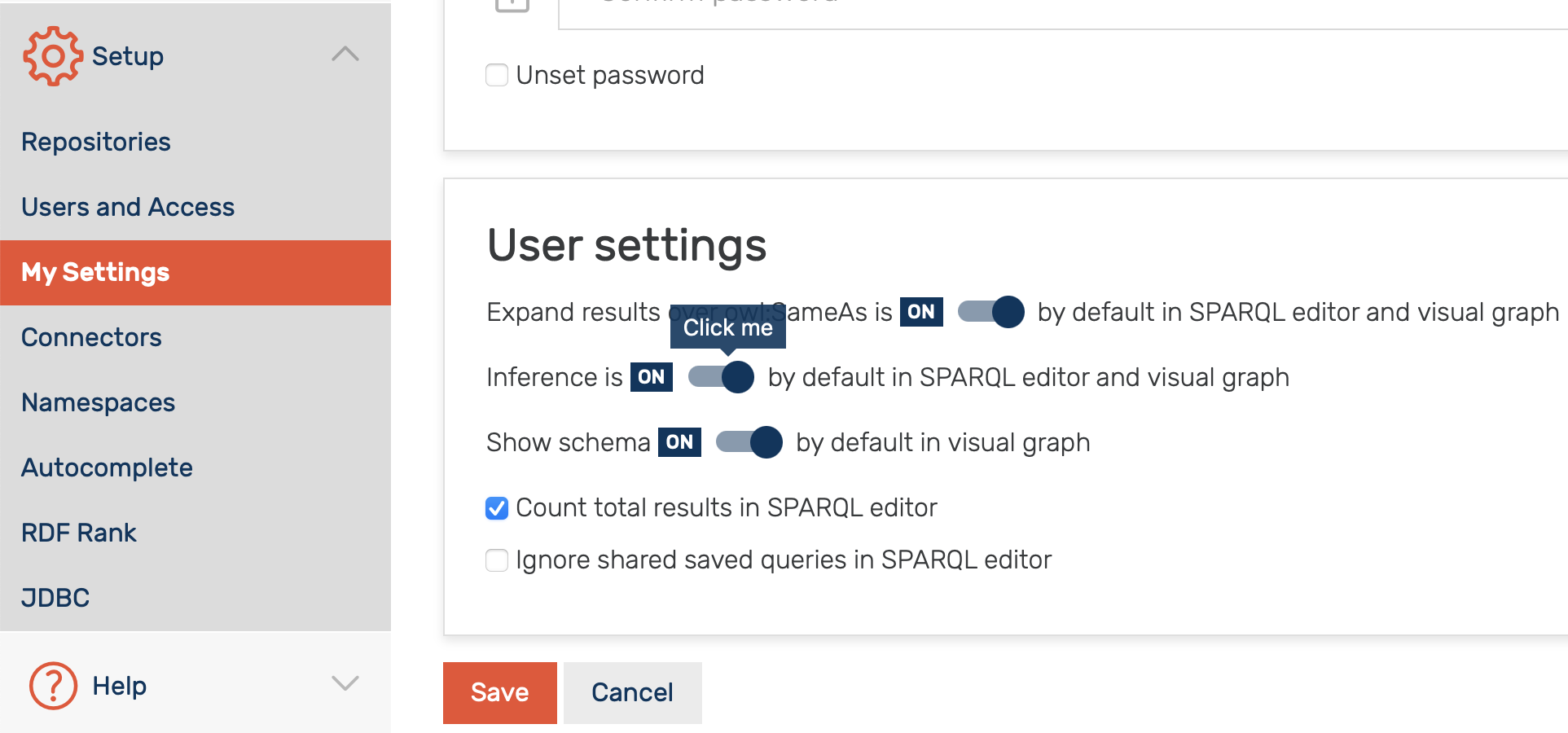
Figure 2: One can choose to exclude/include inferred data in results by selecting the corresponding option in the SPARQL editor settings.
SPARQL Editor
The GraphDB SPARQL editor allows to include or exclude inferred statements in the results by clicking the >> icon, as shown in Figure 3 (see GraphDB documentation for more information).
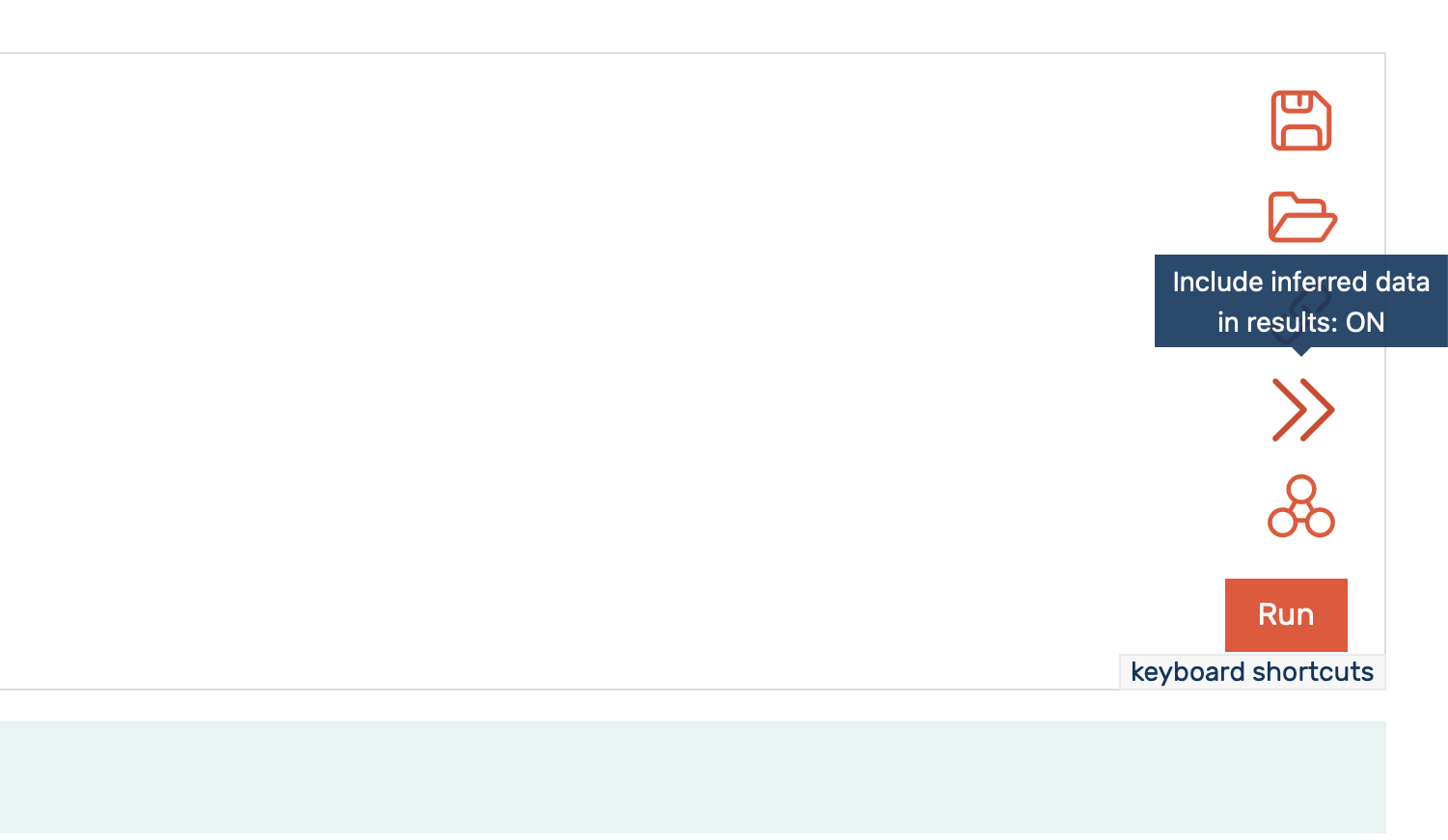
Figure 3: Enabling inclusion (both elements of the arrow icon are a solid line) of inferred statements in the results in GraphDB built in SPARQL editor.
Note
GraphDB System Statements can be used to disable inference from within a SPARQL query, and also to enable it again (assuming the inference was enabled during the repository creation).
Warning
During offline bulk loading of data, no inference is enabled and GraphDB inference settings are ignored (see GraphDB documentation for further info).
SPARQL 1.1
Introduction
SPARQL (SPARQL Protocol and RDF Query Language) is the querying standard semantic language for RDF. More specifically, it is the declarative language part of the W3C standards.
SPARQL queries are based on graph pattern matching finding, meaning that the tool that is doing the search will try to match the given pattern in the query and retrieve the corresponding data.
Shown in Figure 4 is a triple representing a resource resource:HospitalA, which has a relation sphn:hasSubjectPseudoIdentifier to a variable ?patient (note the question mark in front of the variable). This is a valid pattern which can be used for search, and would yield, e.g., the list of patients for resource:HospitalA. The syntax of SPARQL queries is similar to Turtle (but not exactly the same).

Figure 4: Example of a graph.
Structure of a query
A SPARQL query consists of mandatory (bold font) and optional parts:
Prefix declarations, for example:
prefix dc: <...>
prefix uni: <...>
Declare type of query (more information on types of queries):
SELECT
ASK
DESCRIBE
CONSTRUCT
Dataset definition:
FROM <...>
FROM NAMED <...>
Graph pattern (in the form of triples):
WHERE { ... }
Query modifiers:
ORDER BY ...
HAVING ...
GROUP BY ...
LIMIT ...
OFFSET ...
BINDINGS ...
Types of queries
There are four types of queries:
SELECT
get results for requested variables where the output is a table (WC3 documentation)
ASK
check for matches and gives boolean ‘yes/no’ result (WC3 documentation)
CONSTRUCT
get specific parts of a graph, and manipulate graph by creating new triples (WC3 documentation)
DESCRIBE
get basic information about a variable (WC3 documentation)
Query formation
In addition to the already mentioned query types, other constructs are also possible:
Nested queries (i.e., with SPARQL subquery):
one
SELECTinside anotherSELECT(more information)
Federated SPARQL:
query different SPARQL endpoints in the same query using a
SERVICEclause (more information)Note
Some tips for working with SPARQL queries, together with a before/after example:
ais a shortcut forrdf:typePrefixes are highly recommended for better readability
Being familiar with the dataset structure helps to write a query
?patient rdf:type https://www.biomedit.ch/rdf/sphn-ontology/sphn#SubjectPseudoIdentifier ?patient a sphn:SubjectPseudoIdentifier
Query examples
The following examples are based on the mock-data introduced in previous Sections (see mock-data description and loading instructions).
Patients allergic to Peanuts
Here, the question is which patients are allergic to the peanuts. To address this question, the graph pattern to be matched by the query is shown in Figure 5. In this graph, information about the patient is denoted by an instance of a sphn:SubjectPseudoIdentifier class. It is possible to find out which allergy episode links to this patient by matching an instance of an sphn:AllergyEpisode class linked by the sphn:hasSubjectPseudoIdentifier property. Similarly, the substance causing the allergy episode can be found by matching an instance of a sphn:Substance class linked by a sphn:hasSubstance property.
In order to search for patients allergic to Peanuts, the substance can be fixed to Peanut by matching instances of sphn:Substance class linked to snomed:762952008 by the sphn:hasSubstanceCode property.
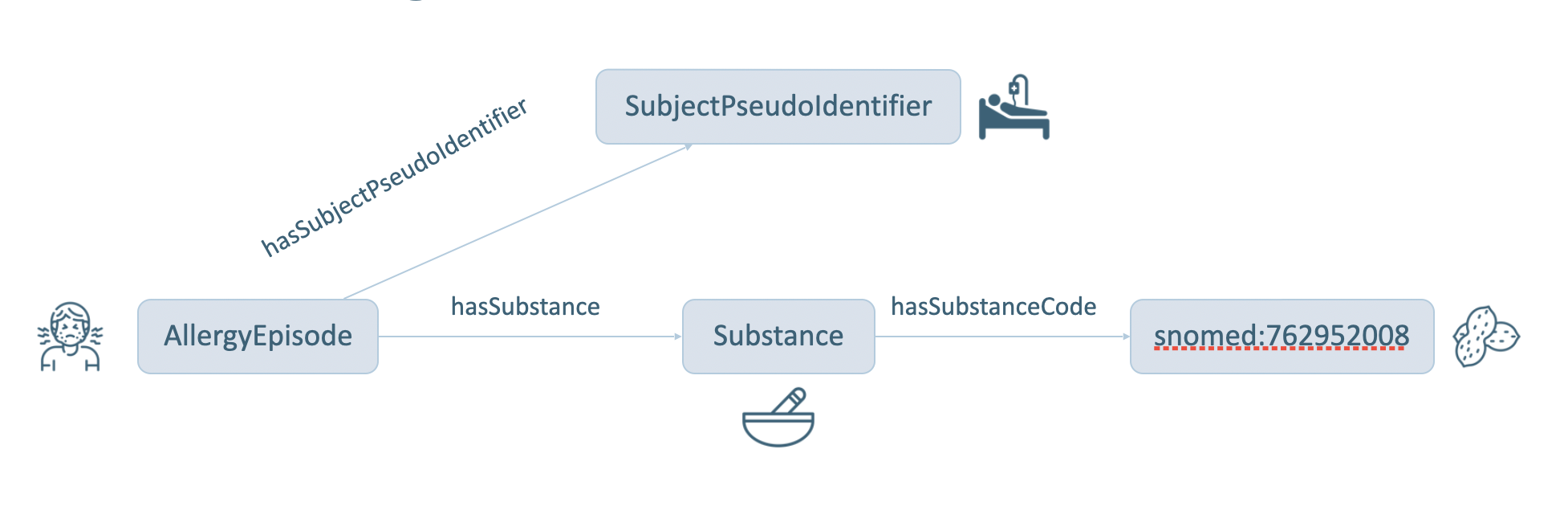
Figure 5: Graph for Patients allergic to Peanuts.
Figure 6 visualizes the graph pattern used for matching Patients allergic to Peanuts, and is followed by the corresponding SPARQL query implementation.
The SPARQL query, after defining the prefixes, retrieves distinct patients (?patient variable of interest). The graph pattern starts by stating that the patients must be of type sphn:SubjectPseudoIdentifier. Next, the ?allergy_episode variable representing the sphn:AllergyEpisode, a ?substance variable representing the sphn:Substance class, and a ?substance_code variable of type snomed:762952008 are defined.
In order to get the data, one still needs to link all of these variables together. To that end, ?allergy_episode is linked to ?patient through the sphn:hasSubjectPseudoIdentifier property, and to ?substance through the sphn:hasSubstance property. Finally, ?substance is linked to ?substance_code through the sphn:hasSubstanceCode property. Running this query in a given triplestore will retrieve any data (i.e., list of patients) matching this graph pattern (see Figure 7 for an excerpt of the results).
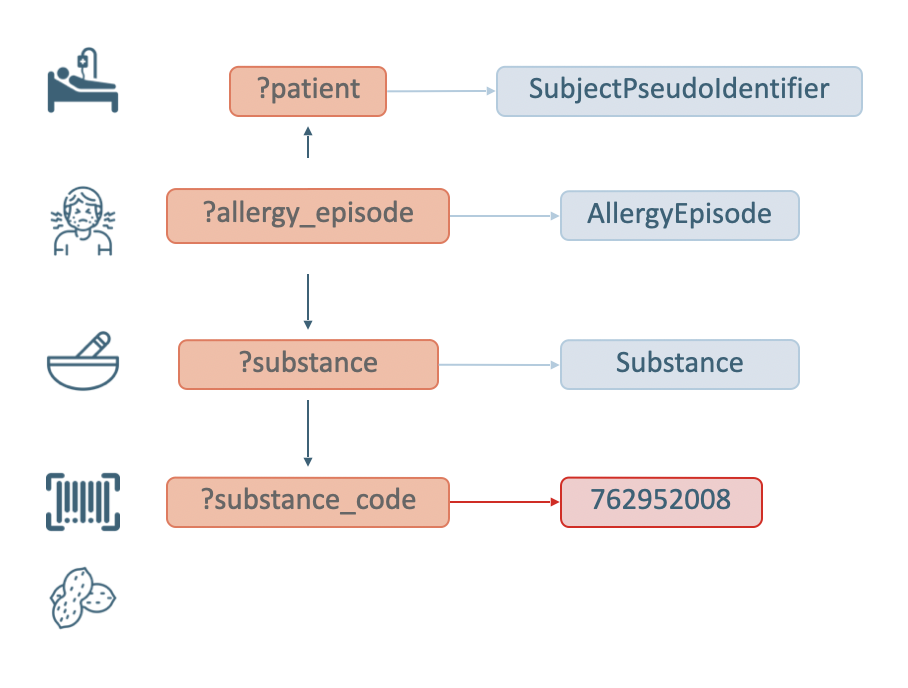
Figure 6: Diagram complementing the SPARQL query for Patients allergic to Peanuts.
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX snomed: <http://snomed.info/id/>
SELECT distinct ?patient
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?allergy_episode a sphn:AllergyEpisode .
?substance a sphn:Substance .
?allergy_episode sphn:hasSubjectPseudoIdentifier ?patient .
?allergy_episode sphn:hasSubstance ?substance .
?substance sphn:hasSubstanceCode ?substance_code .
?substance_code a snomed:762952008 .
}
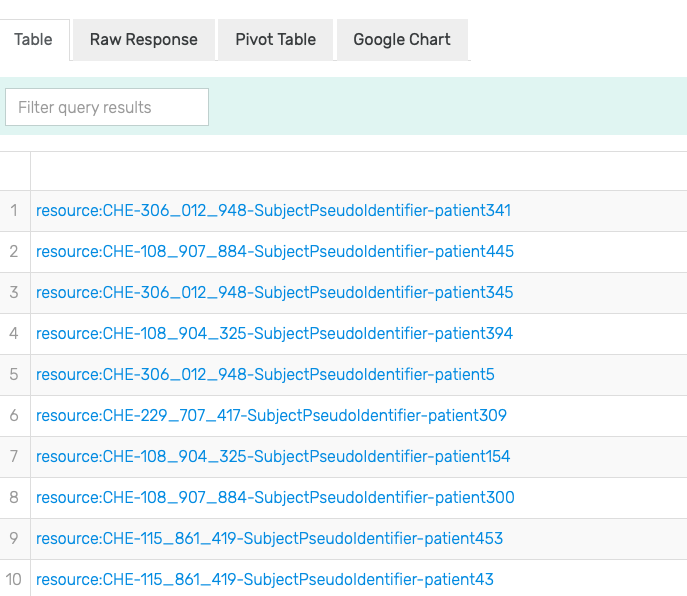
Figure 7: Excerpt of results of running the SPARQL query for Patients allergic to Peanuts in GraphDB on the mock-data.
By modifying the above SPARQL query with the COUNT(distinct ...) statement (see the following code block), it is possible to determine the exact count of the matched patients. For example, evaluating this query in GraphDB with the mock-data used throughout this guide produces the result shown in Figure 8. Note the use of the FILTER(...) statement to retrieve only labels from SNOMED CT.
SELECT (COUNT(distinct ?patient) AS ?patients) ?snomed_code ?label
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?allergy_episode a sphn:AllergyEpisode .
?substance a sphn:Substance .
?allergy_episode sphn:hasSubjectPseudoIdentifier ?patient .
?allergy_episode sphn:hasSubstance ?substance .
?substance sphn:hasSubstanceCode ?substance_code .
?substance_code a snomed:762952008 .
?substance_code rdf:type ?snomed_code .
?snomed_code rdfs:label ?label .
FILTER(strStarts(str(?snomed_code), "http://snomed.info/id/"))
} GROUP BY ?snomed_code ?label
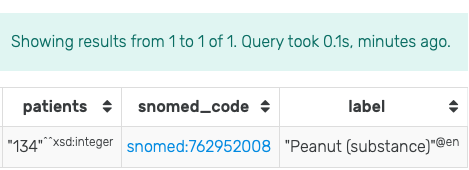
Figure 8: Results of running the modified SPARQL query for Patients allergic to Peanuts in GraphDB on the mock-data.
Patients allergic to Pulse Vegetable
Here, the question is which patients are allergic to pulse vegetable. We do not find this information directly in the data, as the data is often collected at a more granular level e.g. allergy to lentils or beans or beansprouts. Therefore, the hierarchical structure of Pulse Vegetable from SNOMED CT needs to be considered, as shown in Figure 9. However, it is not needed to query individually for all levels in order to get all patients that are allergic to Pulse Vegetable. Thanks to the RDF graph structure and the hierarchy of SNOMED CT, the query can be done in a more straightforward way.
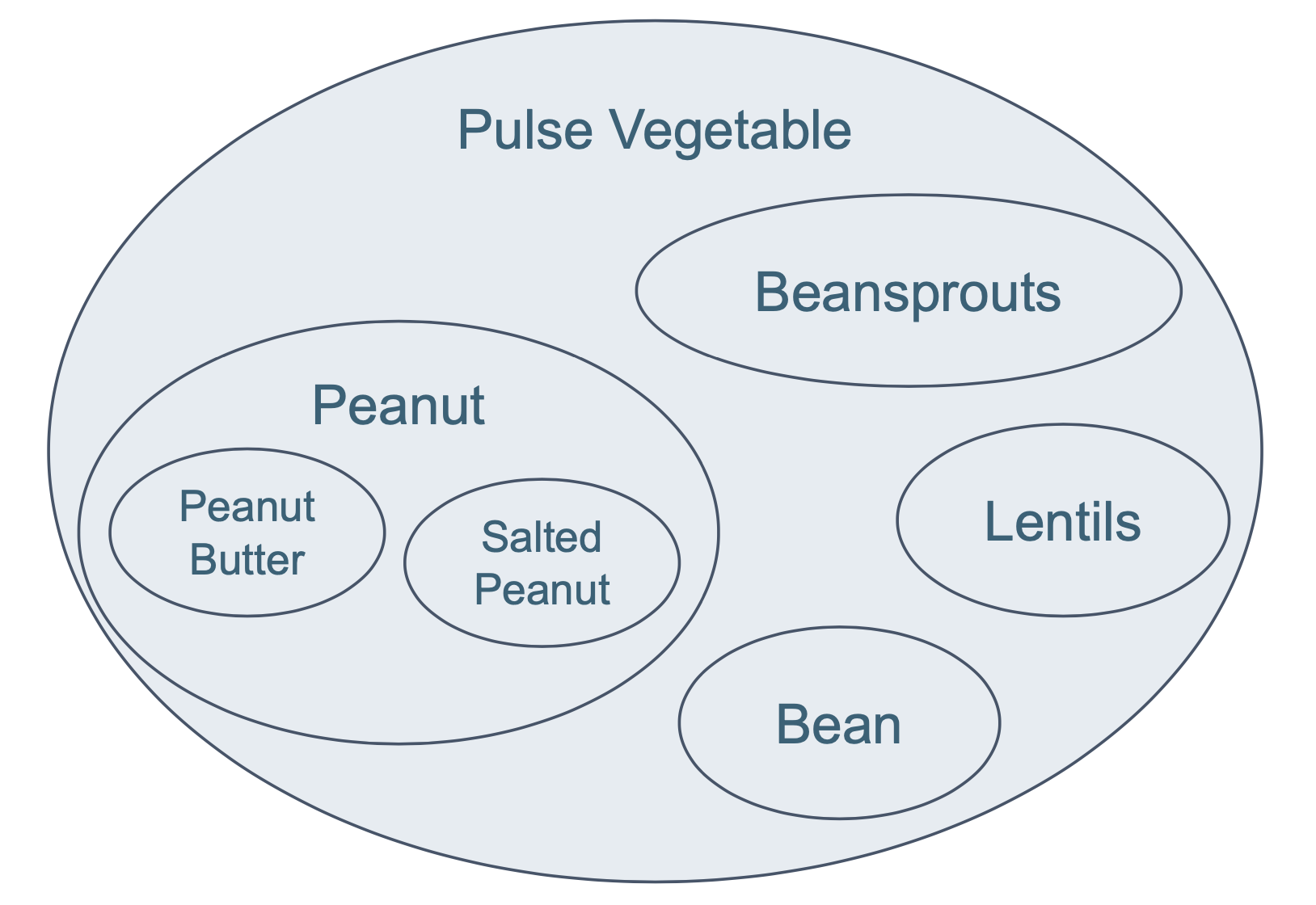
Figure 9: Hierarchical structure of Pulse Vegetable from SNOMED CT.
Query option 1: reasoning without RDF inference
List of patients
Figure 10 visualizes the graph pattern used for matching Patients allergic to Pulse vegetable, and is followed by an implementation enabling reasoning with SPARQL query (Note: without inference turned on).
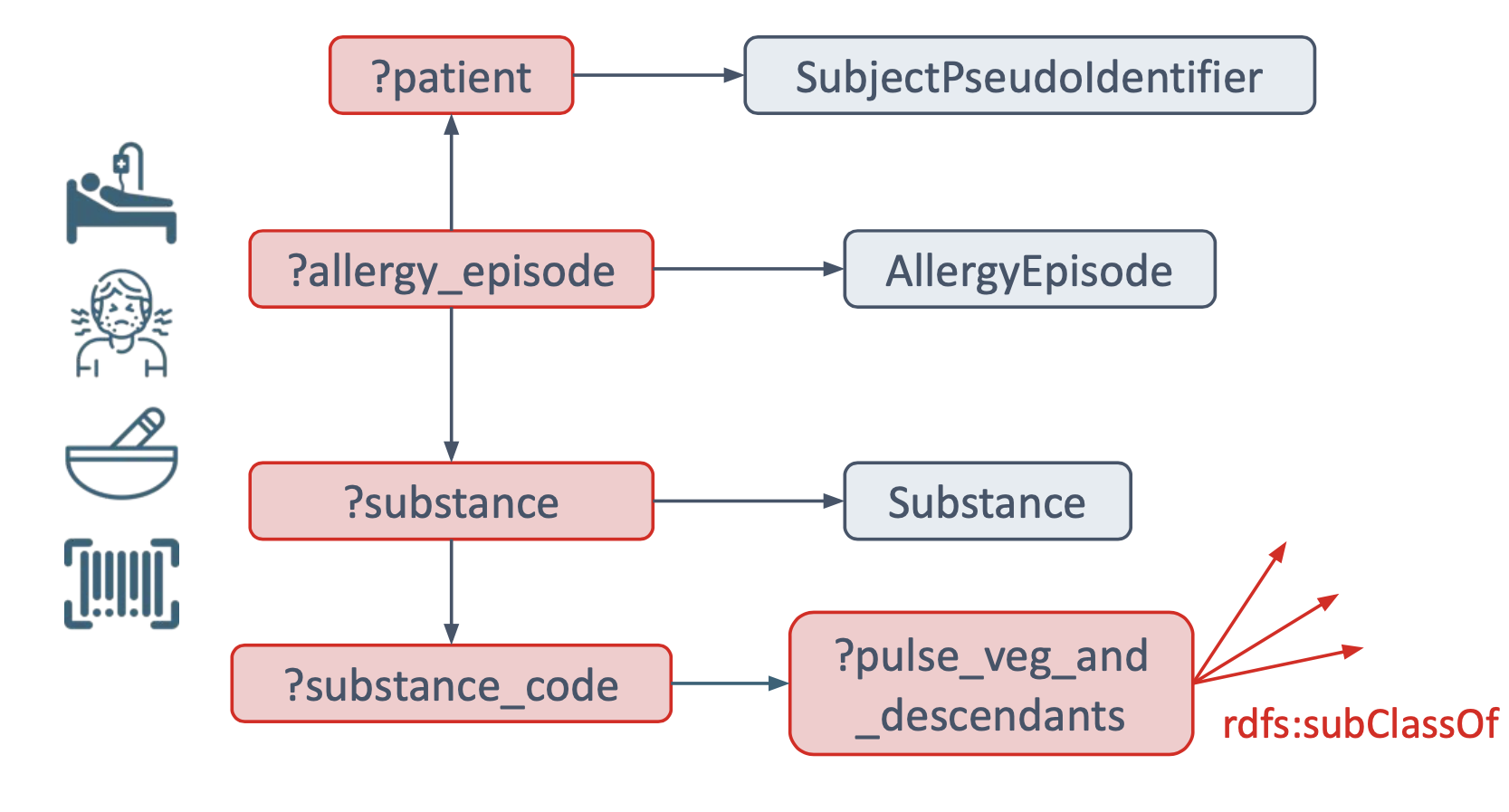
Figure 10: Diagram complementing the SPARQL query for Patients allergic to Pulse Vegetable.
The query, shown in Figure 11 , is in large part, up to and including the row with the ?substance sphn:hasSubstanceCode ?substance_code triple, same as in the previous example in Figure 8. The difference occurs in the statements right after:
the variable
?pulse_veg_and_descendantsis introduced, representing the Pulse Vegetable causing the allergythe
?pulse_veg_and_descendantsis defined as being a subclass of thesnomed:227313005code using therdfs:subClassOf*property. Note that the*following therdfs:subClassOfwill look into all the nested levels from Pulse Vegetable (i.e., not only Beansprouts, Peanut, and other concepts that are immediate subclasses ofsnomed:227313005, but also Peanut Butter, Salted Peanut, etc.).
Running this query in a given triplestore will retrieve any data (i.e., list of patients) matching this graph pattern (see Figure 11 for an excerpt of the results).
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX snomed: <http://snomed.info/id/>
SELECT DISTINCT ?patient
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?allergy_episode a sphn:AllergyEpisode .
?substance a sphn:Substance .
?allergy_episode sphn:hasSubjectPseudoIdentifier ?patient .
?allergy_episode sphn:hasSubstance ?substance .
?substance sphn:hasSubstanceCode ?substance_code .
?substance_code a ?pulse_veg_and_descendants .
?pulse_veg_and_descendants rdfs:subClassOf* snomed:227313005 .
}
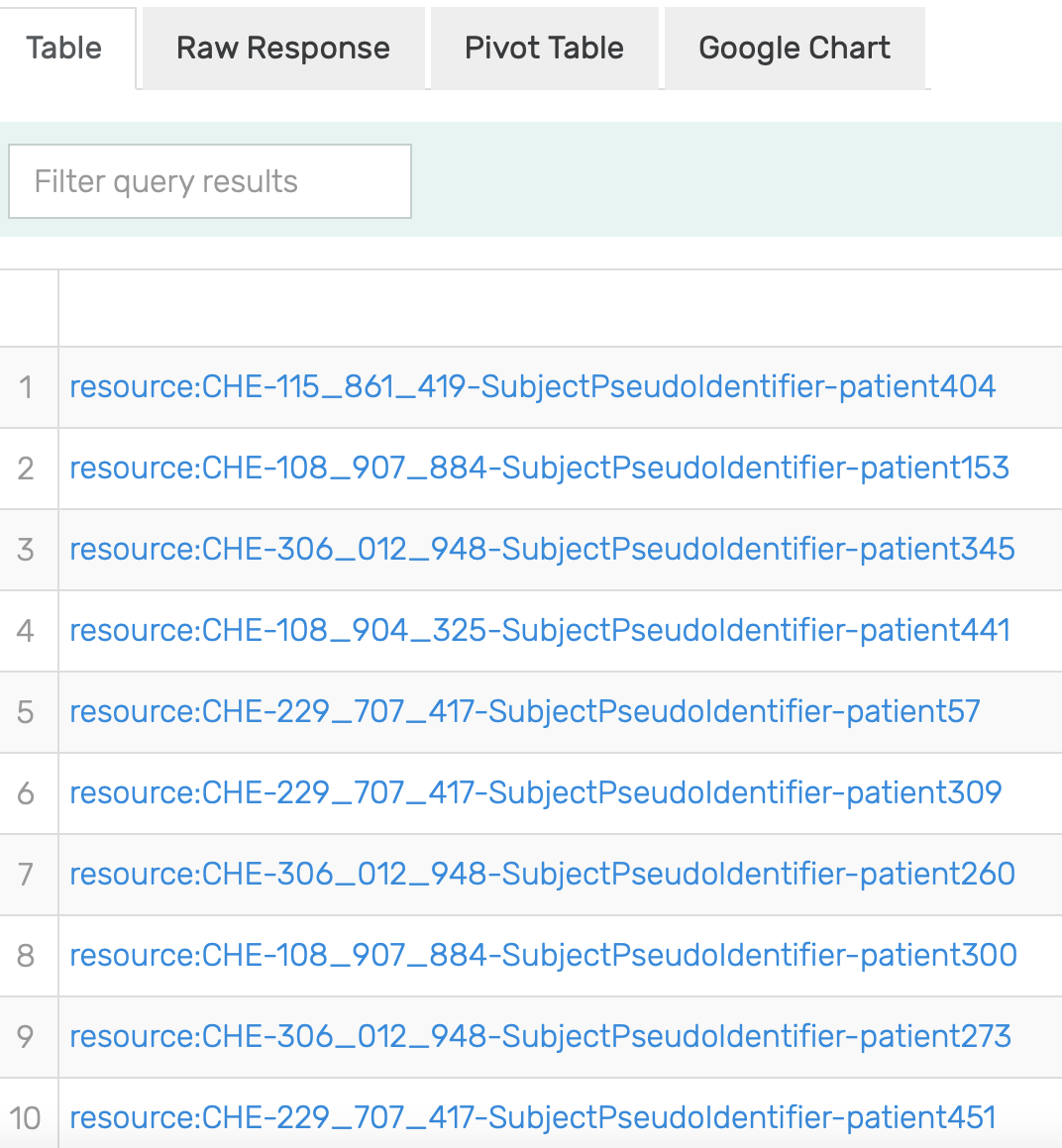
Figure 11: Excerpt of results of running the SPARQL query for Patients allergic to Pulse Vegetable in GraphDB on the mock-data.
Count of patients
By modifying the above SPARQL query with the COUNT(distinct ...) statement (see the following code block), it is possible to determine the exact count of patients allergic to Pulse Vegetable grouped by labels of annotated substance they are allergic to. For example, evaluating this query in GraphDB with the mock-data used throughout this guide produces the result shown in Figure 12. Note the use of the FILTER(...) statement to retrieve only labels from SNOMED CT.
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX snomed: <http://snomed.info/id/>
SELECT (COUNT (DISTINCT ?patient) AS ?patients) ?snomed_code ?label
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?allergy_episode a sphn:AllergyEpisode .
?substance a sphn:Substance .
?allergy_episode sphn:hasSubjectPseudoIdentifier ?patient .
?allergy_episode sphn:hasSubstance ?substance .
?substance sphn:hasSubstanceCode ?substance_code .
?substance_code a ?pulse_veg_and_descendants .
?pulse_veg_and_descendants rdfs:subClassOf* snomed:227313005 .
?substance_code rdf:type ?snomed_code .
?snomed_code rdfs:label ?label .
FILTER(strStarts(str(?snomed_code), "http://snomed.info/id/"))
} GROUP BY ?snomed_code ?label
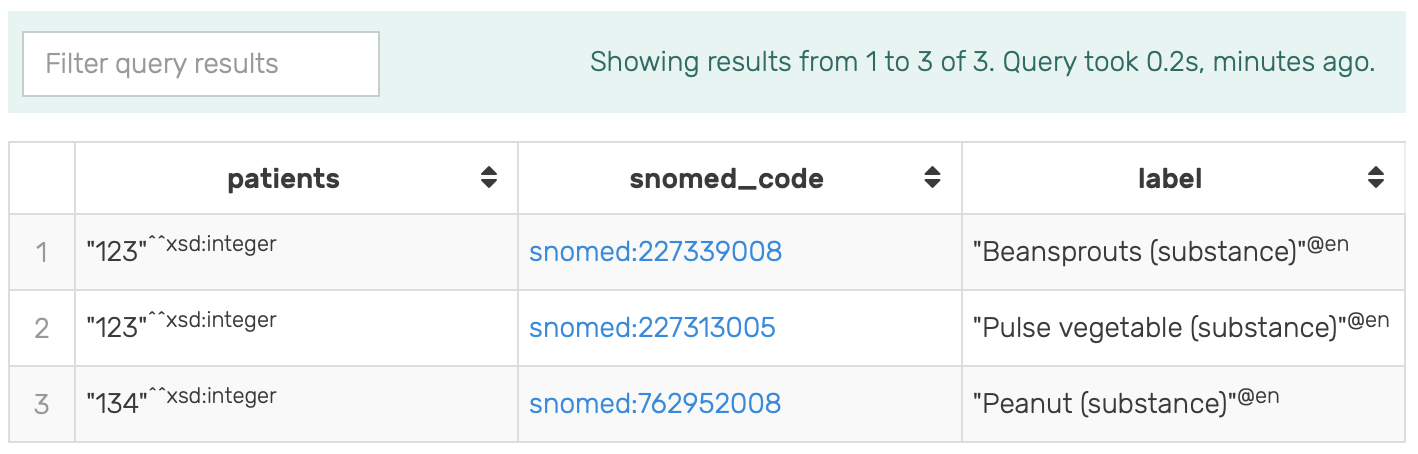
Figure 12: Results of running the modified SPARQL query for Patients allergic to Pulse Vegetable in GraphDB on the mock-data.
RDF reasoning
RDF reasoning enables the computer to infer knowledge based on provided information. For example, given the information in an ontology on hierarchies (Class vs. Subclass; Property vs Subproperty) and in the provided data, the computer is able to do some reasoning with respect to that hierarchy. Two such examples are shown in Figure 13:
based on information coming from the ontology that HeartRate is a Subclass of Measurement, and that data X is a HeartRate, the computer is able to infer that X is a Measurement.
based on information coming from the ontology that hasInsertionSite is a Subproperty of hasBodySite, and that data X hasInsertionSite Arm, the computer is able to infer that X hasBodySite Arm.
In general, patients can have information annotated at different levels of granularity. As mentioned previously, it is not necessary to query individually for all levels of information to get patients that match certain criteria thanks to the RDF graph structure and the hierarchical knowledge provided by SNOMED CT.
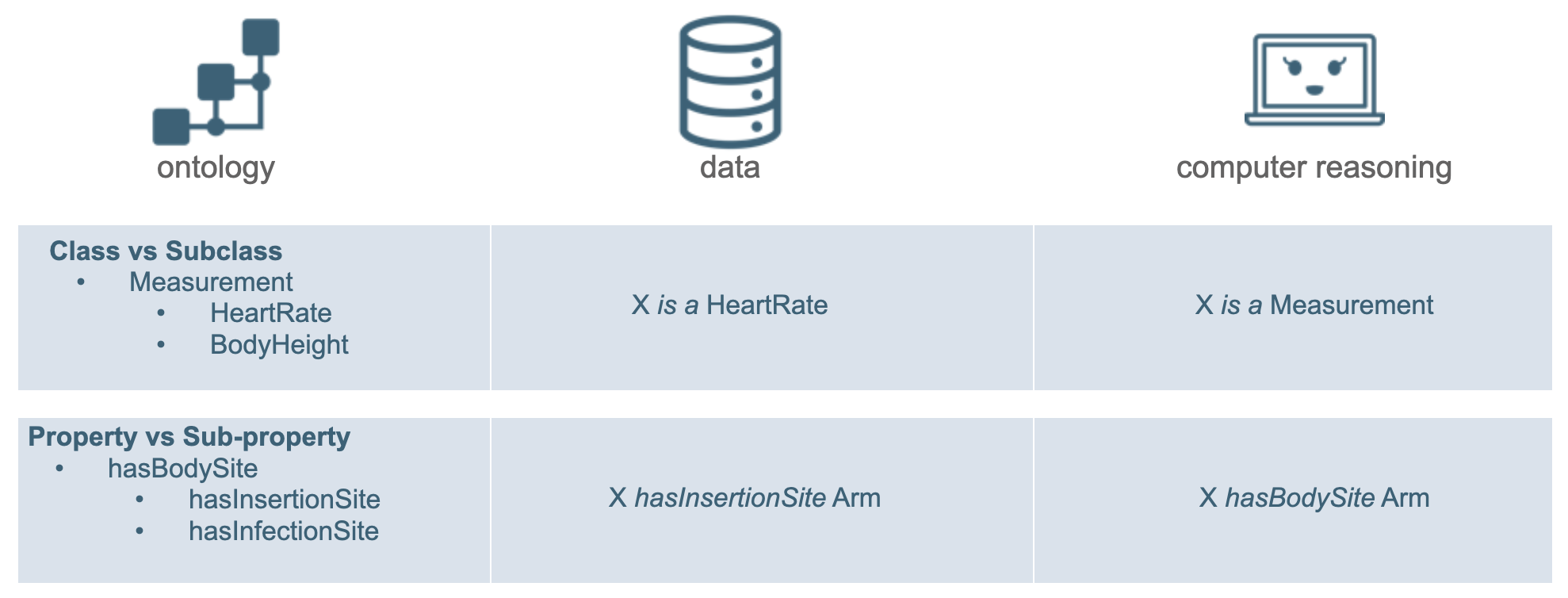
Figure 13: Example of RDF reasoning.
Patients allergic to Pulse Vegetable (inference turned on)
List of patients
Using the reasoning possibilities, the query to retrieve patients allergic to Pulse Vegetable can be simplified. In comparison to when the inference is off, when inference is turned on the main difference occuring in the query statement regarding the pulse vegetables is:
the
?substance_code a snomed:227313005 .line results again in retrieving all the nested levels from Pulse Vegetable. This time, however, without the need of usingrdfs:subClassOf.
Running this query in a given triplestore with inference turned on will retrieve the desired list of patients, as exemplified in Figure 14 (Note the arrows on the right side of the editor that can be used to turn inference ON/OFF).
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX snomed: <http://snomed.info/id/>
SELECT DISTINCT ?patient
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?allergy_episode a sphn:AllergyEpisode .
?substance a sphn:Substance .
?allergy_episode sphn:hasSubjectPseudoIdentifier ?patient .
?allergy_episode sphn:hasSubstance ?substance .
?substance sphn:hasSubstanceCode ?substance_code .
?substance_code a snomed:227313005 .
}
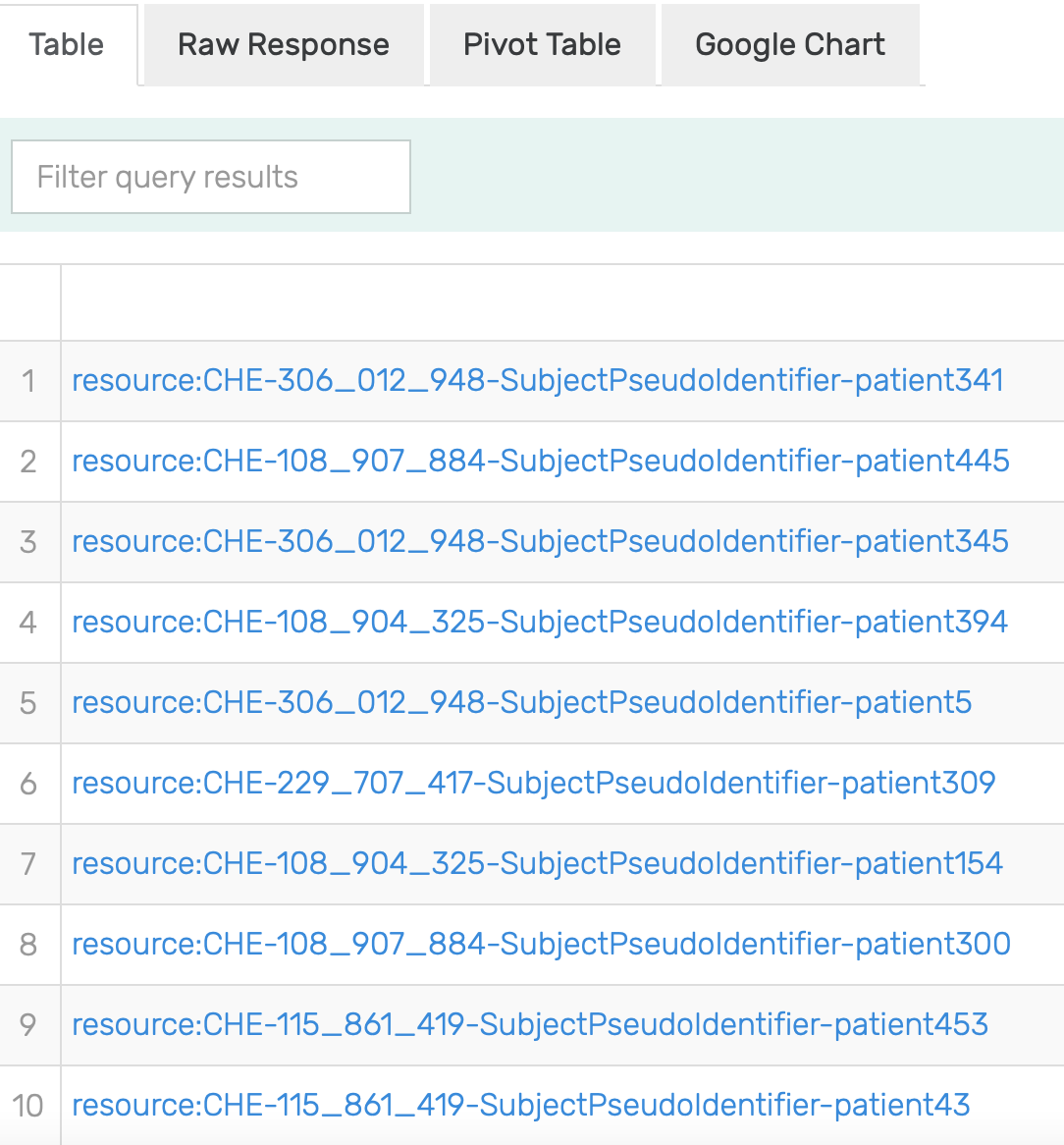
Figure 14: Excerpt of results of running the SPARQL query for Patients allergic to Pulse Vegetable with inference turned on in GraphDB on the mock-data.
Count of patients
By modifying the above SPARQL query with the COUNT(distinct ...) statement (see the following code block) it is possible to determine the exact count of patients allergic to Pulse Vegetable grouped by labels of annotated substance they are allergic to, and with additional inference. For example, evaluating this query in GraphDB with the mock-data used throughout this guide and inference turned on produces the result shown in Figure 15. Note that, with inference turned on, the count of patients allergic to Pulse Vegetable has increased to 380, which is a more accurate statement regarding allergy to any type of Pulse Vegetable.
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX snomed: <http://snomed.info/id/>
SELECT (COUNT (DISTINCT ?patient) as ?patients) ?snomed_code ?label
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?allergy_episode a sphn:AllergyEpisode .
?substance a sphn:Substance .
?allergy_episode sphn:hasSubjectPseudoIdentifier ?patient .
?allergy_episode sphn:hasSubstance ?substance .
?substance sphn:hasSubstanceCode ?substance_code .
?substance_code a snomed:227313005 .
?substance_code rdf:type ?snomed_code .
?snomed_code rdfs:label ?label .
FILTER(strStarts(str(?snomed_code), "http://snomed.info/id/"))
} GROUP BY ?snomed_code ?label
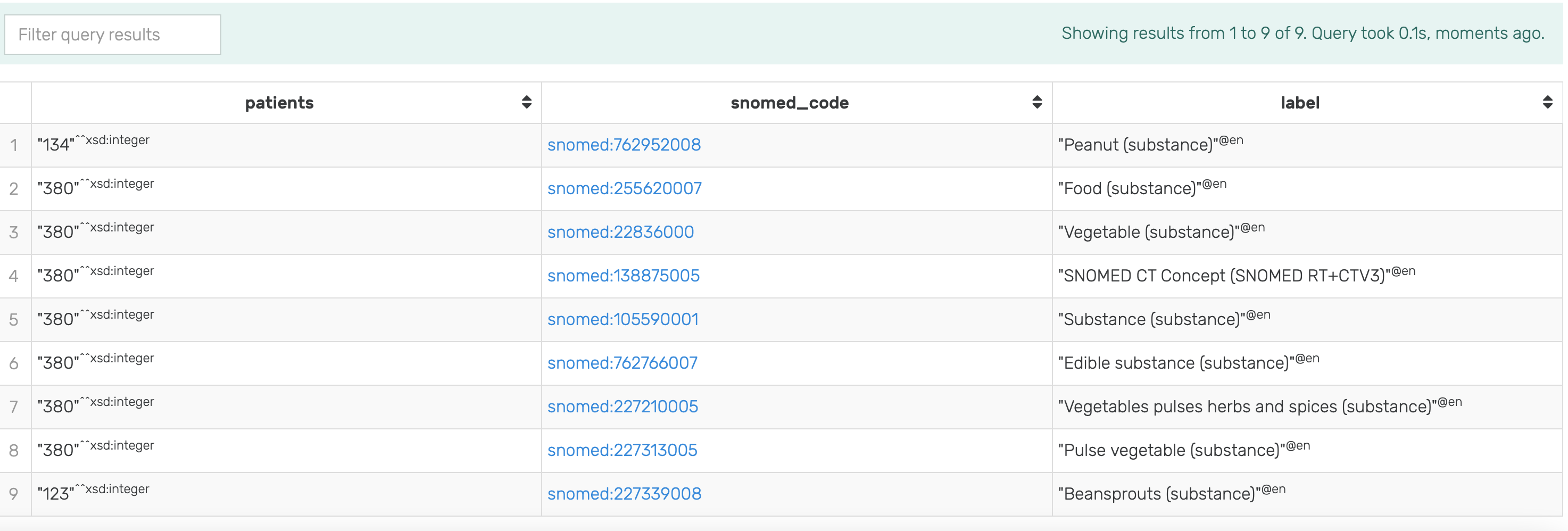
Figure 15: Results of running the modified SPARQL query for Patients allergic to Pulse Vegetable with inference turned on in GraphDB on the mock-data.
Patient with measurements of Leukocytes in Blood by Automated count (LOINC 6690-2)
Here, the question is which patients have had a lab test done identified by a specific LOINC code. To address this question, the graph pattern to be matched by the query is shown in Figure 16. In this graph pattern, instances of a sphn:LabResult class are linked by the sphn:hasSubjectPseudoIdentifier property to instances of a sphn:SubjectPseudoIdentifier class. Patients that had measurements of Leukocytes in Blood by Automated count are queried by matching instances of sphn:LabResult class linked by the sphn:hasLabResultLabTestCode property to loinc:6690-2.
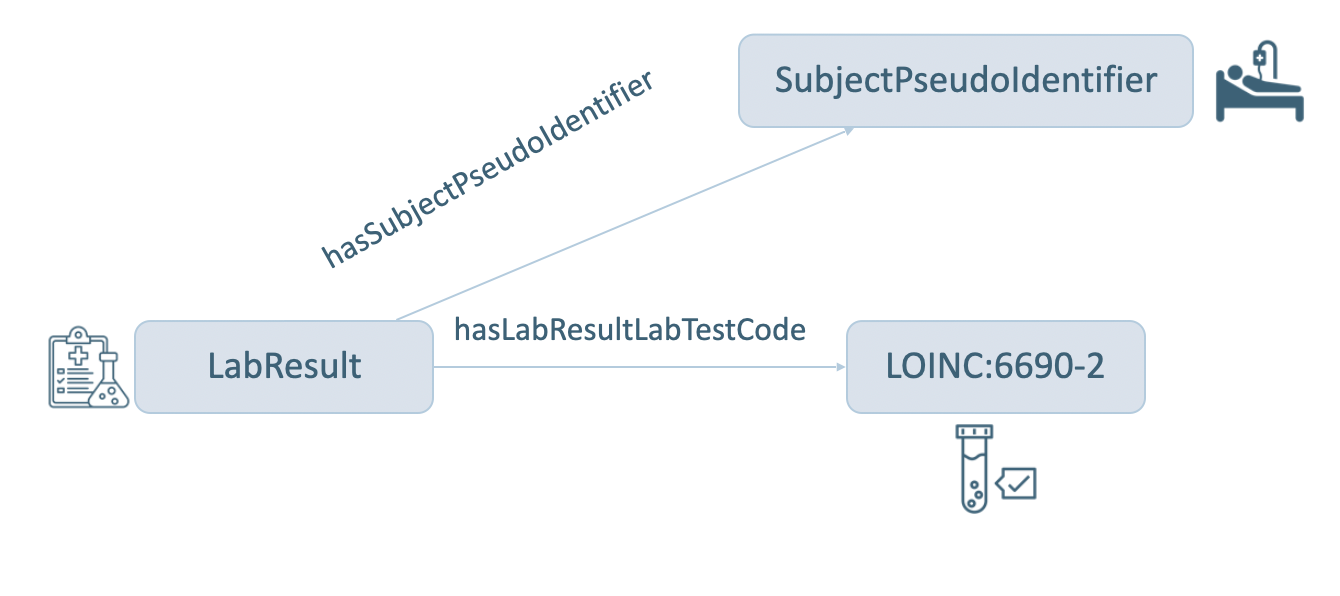
Figure 16: Graph for Patient with measurements of Leukocytes in Blood by Automated count (LOINC 6690-2).
Figure 17 visualizes the graph pattern used for matching Patients with measurements of Leukocytes in Blood by Automated count, and is followed by the corresponding SPARQL query implementation.
Similar as in the previous example, the SPARQL query retrieves distinct patients, and the graph pattern starts by stating that the patients must be of type sphn:SubjectPseudoIdentifier. Next, the ?lab_res variable representing the sphn:LabResult, and a ?test_code variable of type loinc:6690-2 are defined.
In order to get the data, the ?lab_res is linked to ?patient through the sphn:hasSubjectPseudoIdentifier property, and to ?test_code through the sphn:hasLabResultLabTestCode property. Running this query in a given triplestore will retrieve any data (i.e., list of patients) matching this graph pattern. For example, evaluating this query in GraphDB with the mock-data used throughout this guide will retrieve 253 patients annotated with having a lab test code measurements of Leukocytes in Blood by Automated count (see Figure 18 for an excerpt of the results).
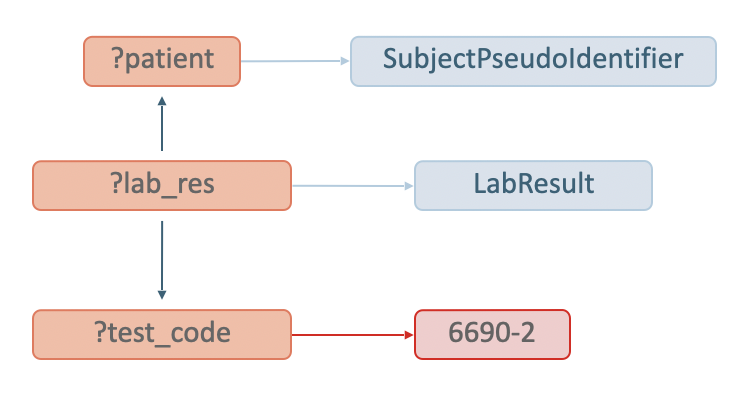
Figure 17: Diagram complementing the SPARQL query for Patient with measurements of Leukocytes in Blood by Automated count.
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX loinc: <https://loinc.org/rdf/>
SELECT distinct ?patient
WHERE {
?patient a sphn:SubjectPseudoIdentifier .
?lab_res a sphn:LabResult .
?lab_res sphn:hasSubjectPseudoIdentifier ?patient .
?lab_res sphn:hasLabResultLabTestCode ?test_code .
?test_code rdf:type loinc:6690-2 .
}
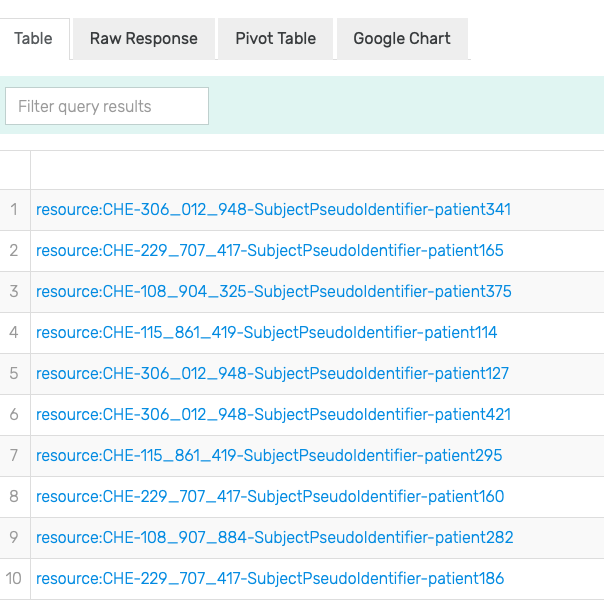
Figure 18: Excerpt of results of running the SPARQL query for Patient with measurements of Leukocytes in Blood by Automated count in GraphDB on the mock-data.
Min/max values for measurements of Leukocytes in Blood by Automated count (LOINC 6690-2)
Here, the question is what are the min/max values measured for a lab test identified by a specific LOINC code. To address this question for measurements of Leukocytes in Blood by Automated count , the previous query can be modified as follow:
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn:<https://biomedit.ch/rdf/sphn-ontology/sphn#>
PREFIX resource:<https://biomedit.ch/rdf/sphn-resource/>
PREFIX xsd:<http://www.w3.org/2001/XMLSchema#>
PREFIX loinc: <https://loinc.org/rdf/>
PREFIX sphn-loinc: <https://biomedit.ch/rdf/sphn-resource/loinc/>
SELECT ?test_code (MIN(?lab_res_value) AS ?min_value) (MAX(?lab_res_value) AS ?max_value) ?lab_res_unit
WHERE {
?lab_res a sphn:LabResult .
?lab_res sphn:hasLabResultLabTestCode ?test_code .
?test_code rdf:type loinc:6690-2 .
?lab_res sphn:hasLabResultValue ?lab_res_value .
?lab_res sphn:hasLabResultUnit ?lab_res_unit.
}
GROUP BY ?test_code ?lab_res_unit
Running this query in GraphDB with the mock-data used throughout this guide produces the result shown in Figure 19. Note the use of the GROUP BY ... statement to group the retrieved results.
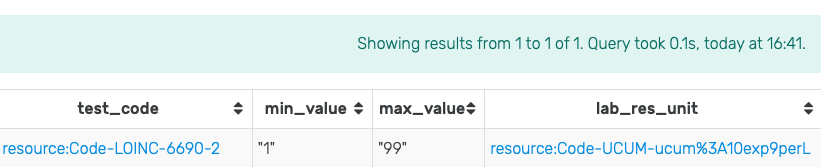
Figure 19: Results of running the SPARQL query for min/max values for measurements of Leukocytes in Blood by Automated count (LOINC 6690-2) in GraphDB on the mock-data.
References
Further information is available in the following references: