USZ RDF Pipeline early version based on CSharp
Note
The following documentation about implementation is provided by Universitätsspital Zürich (USZ). For additional information or questions, please send an email to SPHN Data Coordination Center at dcc@sib.swiss.
Overview
This project was initiated by the HospIT WG Taskforce for Data Exchange and was developped as an early USZ specific RDF pipeline.
With the RDF Extract the research data can be exported in a graph respresentation and it uses RDF as the underlying exchange format.
In April/May 2021 the RDF Schema 2021.1 based on the SPHN ontology 2021.1 was created. The current version v2 supports this 2021.1 release. Not all future changes in the RDF Schema will need a new version of the RDF extract.
The creation of RDF files is based on the following 7 steps:
Requirements for RDF Data Extractions
Create the project data mart
De-Identify the data in the data mart
Map data in the data mart to target value sets
Create standardized RDF (SPHN) views
The RDF Ontology and Mapping
RDF Extraction into a turtle file
RDF Validation
In order to process the source data the data engineer has to create views based on the view specification for each concept which in turn is based on the SPHN or project ontology and, if available, the SPHN or project specific documentation in Excel. Steps 1-4 are part of the View creation.
The views are structured to contain all the sub-concepts of a concept. With this we can assure that all the referenced RDF concept are also instantiated. The RDF files will be created by patient and as a consequence some concepts like Unit will have multiple instantiation, however, while loading this data into a graph these redundand concept instances will be merged.
The extract process has different option but it will generally generate one RDF file per patient for a given project depending on which concepts the project requires and that are set to active in the Projec Class config table.
Once an RDF file is generated it’s structure and values can be checked with the quality control functionality which is provided by SPHN. This QC functionality can help validate the RDF extract but at it’s current stage it is difficult to take actions on the reported issues. This is why DCC and the RDF expert group are investigating better and more streamlined options.
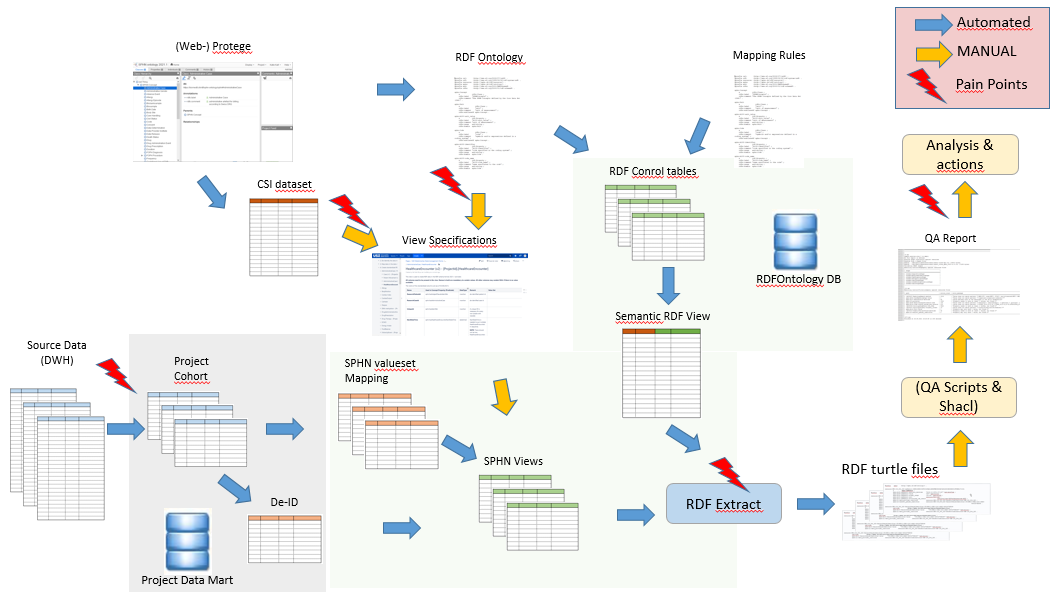
0. Requirements for RDF Data Extractions
In order to keep the RDF extract solution simple, it is necessary to follow the below rules to prepare the project data.

1. Create the project data mart
Preparation of data mart tables
The data mart is a project specific database that contains all the data that needs to be represented in the input views (see step 4) for RDF export.
In order to simplify the creation of the RDF Views, it is adviced to structure the project data mart along side the RDF views.
The views currently supported are listed in chapter 4. Project specific enhancements to SPHN concepts are merged into the SPHN view definition and marked specific for which project they are required. Project specific enhancements need only be provided for the specific project and can be omitted by other projects.
Value Sets
Before the preparation of the data mart it should be defined how the value sets for the variables are to be delivered. At the moment the assumption is that the RDF Views map the value sets to the target value sets (currently SPHN). The mapping rules are described in Step 3. In order to reduce efforts for mappings, the value set has to be defined at the beginning and should not be changed during the development of the data mart as it would cause extra maintenance of the mapping rules.
2. De-Identify the data in the data mart
As the RDF extraction only extracts the data the de-identification has to be performed before hand. How the de-identification is to be performed is specified in a separate de-identification concept.
In order to have a simple process and for troubleshooting the identifying data doesn’t need to be removed from the data mart unless some users will have access to the data mart too. If both are present, the de-identified data needs to be clearly marked.
3. Map data in the data mart to target value sets
As of March 2022 the following mapping tables are defined.
AdministrationRoute
AdministrativeGender
BodySite
CareHandlingType
CivilStatus
DeathStatus
DiagnosisRank
DrugCode
FollowUpEvent
Galenic
GeneCode
GeneName
Intention
LaboratoryCode
Laterality
LocationClass
MaterialTypeLiquid
MicroorganismCode
MicroorganismKingdom
MicroscopyCellType
MicroscopyMorphology
ProcedureRank
ProviderInstitute
Quantity
RadiologyMethods
RadiotherapyProcedure
RadiotherapyProcedure
SampleSource
Score
StainingResult
TestMethod
TherapeuticArea
TumorGrade
TumorStage
Unit
VentilationDevice
VentilationMode
Each of the mapping table is stored in the DB Schema [MAP] and its basic structures are not standardized and they use as a minimum the following columns:
USZ_Value (source value from the CDP or source system)
SPHN_Value (that corresponds to SPHN value set defined as individuals)
SNOMED_Value (default value sets that are expressed in Snomed CT)
LOINC_Value (For LabResultCode mappings or where coding is available in LOINC)
UCUM_Value (For units mapping)
Since RDF schema version 2021.1 value sets that are defined in the ontology are available in table [RDF].[AttributeValueSet] for cross validation or the target mapping codes. Limitation is where the ontology just specifies the value set as child of for Snomed CT.
4. Create standardized RDF (SPHN) views
In order to have a stable mapping definition between RDF and the source data it is crucial to have a standard definition of views with standardized names. These views are stored in the RDFOntology database.
The schema name has to correspond to the project code as defined in the RDFOntology database.
Example View Definition Patient (v3) - [ProjectId].[Patient]
This view is used to create RDF data in the RDF schema format 2022.1 and later.
This view contains the data to create the following concepts instantiation (please refer also to https://sphn-semantic-framework.readthedocs.io/en/latest/user_guide/data_generation.html):
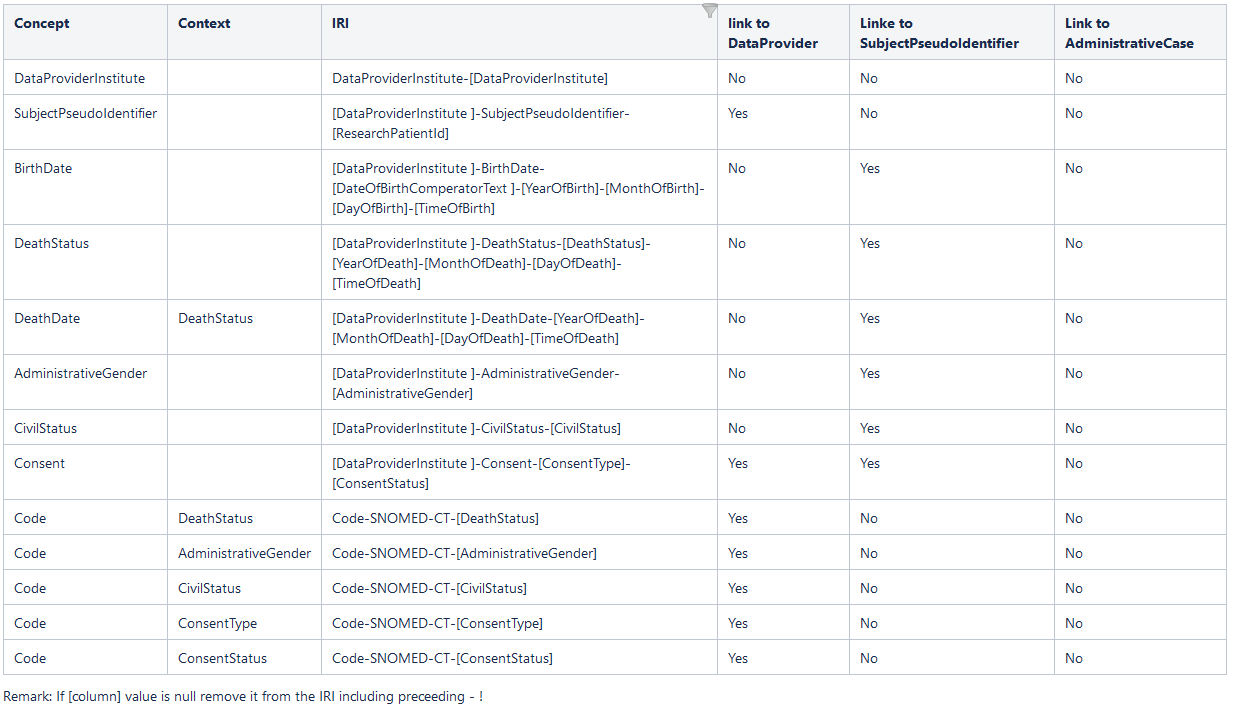
Remark: If [column] value is null remove it from the IRI including preceeding - !
Note
All columns need to be present in the view.
Names in bold are mandatory and must contain non-null values.
All other columns may contain NULL if there is no value available.
Lines marked in BLUE have changed versus the old version.
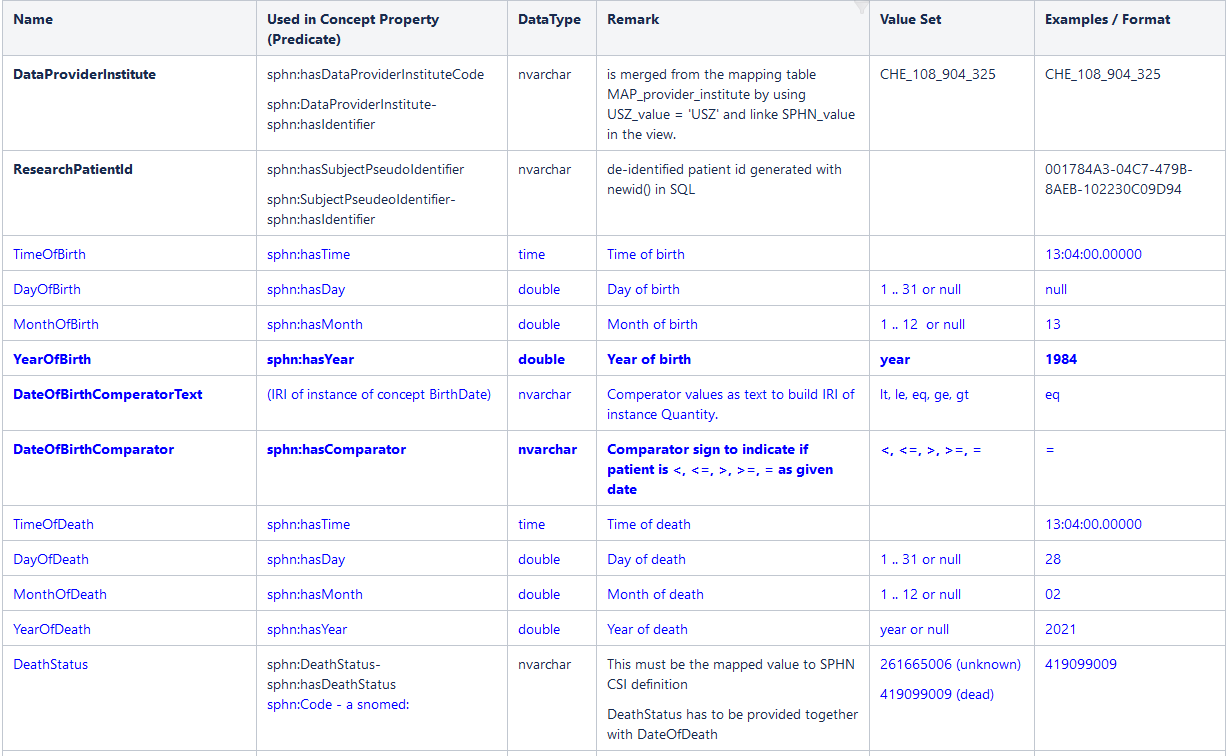
Excerpt of the standardized view for patients as of 01.06.2022 as an example (not all row shown):
5. The RDF Ontology and Mapping
RDF Ontology (RDF Schema 2021.1)
Introduction
The ontology is in RDF turtle format and consists of several sections which are described below.
Note
In the examples below every line that starts with one or more # denotes a comment line!
Prefixes (Header section)
List of prefixes with their corresponding URI’s. The prefixes are used throughout the ontology to substitute the full URI and make the ontology more readable.
Every prefix used in the ontology needs to be defined here in the format:
@prefix [prefix-value]: [URL].
Metadata Section
Every ontology needs to have a metadata node that defines some basic properties about the ontology. These are:
owl:versionIRI- This is defining the version of the ontology and corresponds to the version of the SPHN or project data set. It is usually in line with the RDF schema version, however, this does not need to be the case.
owl:imports- Lists all external taxonomies that this ontology works with. It is relevant in regards of the validation of the valuesets specified in the ontology (see below).
dct:license- Every ontology from SPHN is distributed based on the creativecommons license which is defined here.
dc:descriptionand dc:title - These are just additional names to describe the ontology.
The metadata is not used for generating RDF data at the moment.
Annotation Properties Section
Some of the properties in the ontology are described by non-RDF descriptors and in order to use them these need to be defined in the ontology. They are all of rdf:type owl:AnnotationProperty. These annotation definitions are not used to generate the RDF extract. They are necessary if the ontology will be loaded into the RDF framework like rdflib (Python) or dotnetrdf (C#).
### http://purl.org/dc/terms/license
dct:license rdf:type owl:AnnotationProperty .
### http://www.w3.org/2002/07/owl#oneOf
owl:oneOf rdf:type owl:AnnotationProperty .
Object Properties Section
RDF differentiates between object properties and data properties (see next section). Object properties represent the edges of a graph or in other words the link between different nodes. As a consequence the opject property always generate an IRI object.
Every object property is defined by the following predicates:
rdf:type- The type for object properties is always ObjectProperty
rdfs:subPropertyOf- all the properties of an ontology are subclasses of other properties or the root properties sphn:SPHNAttributeObject or a corresponding project specific root property like psss:PSSSAttributeObject.
rdfs:domain- This predicate defines for which RDF class the property can be used. Since RDF schema version 2021.1 this can be a one to one relation like in the sphn:hasCareHandlingTypeCode example below or it can be used for several different classes as shown in the example under sphn:hasAdministrationRouteCode or sphn:hasCode. In the later case it is a construct that is defined over several lines and uses the owl:unionOf predicate.
rdfs:range- This predicate defines where the property can point to via the IRI. All the values of the range are classes itself. As with rdfs:domain the rdfs:range can also be a one to one (sphn:hasAdministratonRouteCode below) or a one to many relation (sphn:hasCareHandlingTypeCode). Again the one to many type is defined over several lines with the owlunionOf predicate. The difficulty with the rdfs:range is that it is not always clear if it’s a one to one or not. In the example sphn:hasAdministratonRouteCode below it states only one snomed code as range but in actual fact it is a root element from which all the sub-elements can be pointed to (child of SCT:284009009) in the Excel documentation. Additionally the entry under range can also point to a valueset class as is shown in example sphn:hasDeathStatus below, however, by the _ sign in the range value it can be deferred that the Death_status is actually a valueset defined by SPHN and the value Death_status is defined somewhere in the ontology as a rdfs:subPropertyOf sphn:valueset. To make the whole definition more complex, the property can actually also point to an instance of a class that is defined under the superProperty (the one mentioned under rdfs:subPropertyOf) under its rdfs:range.
rdfs:isDefinedBy- This predicate represents the meaning binding of a property and it lists the IRI of external taxonomies like snomed or loinc with the corresponding code that defines this property. This information is not used to generate the RDF extract.
rdfs:commentandrdfs:label- These predicates are used to describe the property in addition to rdfs:isDefinedBy thatis not always present.
Data Properties Section
RDF differentiates between object properties (see section above) and data properties. Data properties represent the data describing a node of a graph. Therefore the data property always is represented with an object (triple) that is a datatype.
Every data property is defined by the following predicates:
rdf:type- The type for data properties is always DatatypeProperty
rdfs:subPropertyOf- all the properties of an ontology are subclasses of other properties or the root properties sphn:SPHNAttributeDatatype or a corresponding project specific root property like psss:PSSSAttributeDatatype.
rdfs:domain- This predicate defines for which RDF class the property can be used. Since RDF schema version 2021.1 this can be a one to one relation like in the sphn:hasAdministrativeGenderDateTime example below or it can be used for several different classes as shown in the example under sphn:hasEndDateTime. In the later case it is a construct that is defined over several lines and uses the owl:unionOf predicate.
rdfs:range- This predicate defines the data type of the object. All the values of the range are URIs to xsd datatypes like xsd:string or xsd:dateTime or xsd:double or xsd:gYear..
rdfs:commentandrdfs:label- These predicates are used to describe the property in addition to rdfs:isDefinedBy thatis not always present.
Class Section
Classes in RDF correspond to the concept it describes.
Every Class is defined by the following predicates:
rdf:type- The type for data properties is always owl:Class
rdfs:subClassOf- all the properties of an ontology are subclasses of other properties or the root properties sphn:SPHNConcept or a corresponding project specific root property like psss:PSSSConcept.
rdfs:commentandrdfs:label- These predicates are used to describe the property in and give a readable Name.
owl:equivalentClass- For classes the rdfs:isDefinedBy is not used and instead a separate owl:equivalentClass is used as shown in the example below. The definition is thougth the other way round where the defining object is represented as the triple-subject and the triple-object represents the class name. This definition is currently not parsed while loading the ontology into RDFOntology database.
Namedindividuals Section
A named individual is an object that represents a value from a valueset. These value set definitions are introduced in the RDF 2021.1 schema and can be used to validate data against allowed value sets but they can also be used to create mapping tables.
Currently they are loaded into the RDFOntology database but are not used for any processing.
Every Namedindividual is defined by the following predicates:
rdf:type- The type for Namedindividual is always defined by owl:NamedIndifidual and sphn:{Valueset_name} where as valueset_name always refers to an RDF Class that combines all values of a valueset into a class definition (see section below).
rdfs:label- This predicates is used to give the value set a more readable name.
SPHN Valueset Definitions
The RDF Schema 2021.1 does offer two types of valueset definitions. The first one is a valueset defined by SPHN or a project with a finit list of values. These will be linked in the object properties (IRI) that used them with the sphn or project prefix and value (i.e. sphn:AmnioticFluid).
In the Ontology you will finde for these valueset definitions the following three informations:
ObjectPropertyrefers to a valueset definition class in the predicate rdfs:range where the range is a class defined as rdfs:subClassOf sphn:ValueSet and the name usually consist of the Class name of the property that uses it plus a valueset name.A
Valueset Classof type rdfs:subClassOf sphn:ValueSet with a predicate equivalentClass that is of rdf:type owl:Class and lists all the possible values as SPHN or project prefix : value.A
namedindividual, as described in the section above, for each allowed value.
Standard Taxonomy Valueset Definitions
The RDF Schema 2021.1 does offer two types of valueset definitions. The second one is a valueset defined by external taxonomies like snomed, Loinc, atc etc.
This type of value set doesn’t need any additional definitions in the ontology and can directly be specified in the range of the ObjectProperty where it applies.
There are usually two different ways how they are specified:
The range of the ObjectProperty lists a range of Classes (IRI’s) that refer to a value in the external ontology. In this case any value specified in the list can be used as a valid value. Sometimes also values that are a child of the value specified in the range can be used. This is not necessary visible in the ontology and is part of the specification (Excel). The values in the range do not have to belong to the same external taxonomy, they can even be mixed with another class that is not a value set (see last example below for Code, atc and snomed).
There is only one value specified in the range of the ObjectProperty and this means that all child elements of this value are acceptable values in the valueset. This is usually used for SNOMED-CT.
Defining RDF ontology mappings (2021.1)
Overview
This section documents how to create RDF mapping rules based on the example of the concept “FOPHProcedure”.
Note
Please keep in mind that creating the mapping rules is a fragile process at the moment and it is important to follow this procedure exactly in order to prevent any issues in the extraction process!
Workspace setup
In order to load the ontology and mapping the input file (turtle) for the load process needs to be a complete graph. This means that in order to load mapping rules for a class or property the underlying ontology definition of the class or property needs to be in the same file. To prevent any issues and keep the ontology and mapping file readable it needs to have the following structure:
SPHN prefix definition
Project specific prefix definitions (optinal)
USZ prefix definitions for the mapping rules
SPHN concepts/classes and properties
Project specific concepts/classes and properties (optional)
USZ ontology definitions for the mappings
SPHN mapping rules
Project specific mapping rules
In order to achieve this following directory anf partial file structure is proposed:
SPHN/
SPHN_Prefix.ttl - The prefix definitions as stated in the SPHN ontology.
SPHN_Ontology.ttl - The SPHN ontology definition (part after prefix definition)
SPHN_Mapping.ttl - The SPHN mappings
{ProjectId}/
{ProjectId}_Prefix.ttl - The project specific prefix definitions as stated in the project ontology.
{ProjectId}_Ontology.ttl - The project ontology definition (part after prefix definition)
{ProjectId}_Mapping.ttl - The project mappings
USZ/
USZ_Prefix.ttl - The prefixes defined by USZ for the mapping rules
USZ_Ontology.ttl - USZ annotation ontology definition used for the mapping rules
After all the files are created and the mappings defined the following PowerShell script will create a file {projectId}_ontology_mapping.ttl. This fill will later be used to load the mapping rules into the RDFOntology database.
Please change the ProjectID accordingly as well as the path definition. It is suggested to only change the root path and let the structure of the other sub-directories be as defined in the script.
cd D:\KTK\RDFData\
rm {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl
Get-Content SPHN\Ontology\v2021.1\SPHN_prefix.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content {ProjectID}\Ontology\v2021.1\{ProjectID}_prefix.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content USZ\Ontology\v2021.1\USZ_prefix.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content SPHN\Ontology\v2021.1\SPHN_ontology.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content USZ\Ontology\v2021.1\USZ_ontology.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content SPHN\Ontology\v2021.1\SPHN_mapping.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
Get-Content {ProjectID}\Ontology\v2021.1\{ProjectID}_mapping.ttl | Out-File -Append {ProjectID}\Ontology\v2021.1\{ProjectID}_ontology_mapping.ttl -Encoding ascii
USZ Prefix and Annotations
For creating the mapping the following prefix definitions are required:
@prefix usz: <http://www.usz.ch/rdf/ontology/> .
@prefix uszm: <http://www.usz.ch/rdf/ontology/mapping#> .
The URL for the prefixes doesn’t have any special meaning at the moment as we do not have any web-service/api that uses them at the moment.
The annotation definition required are:
#################################################################
# Annotation properties
#################################################################
### http://www.usz.ch/rdf/ontology/mapping#ColumnName
uszm:ColumnName rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:ColumnName" .
### http://www.usz.ch/rdf/ontology/mapping#Context
uszm:Context rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:Context" .
### http://www.usz.ch/rdf/ontology/mapping#DatabaseName
uszm:DatabaseName rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:DatabaseName" .
### http://www.usz.ch/rdf/ontology/mapping#Filter
uszm:Filter rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:Filter" .
### http://www.usz.ch/rdf/ontology/mapping#HashKey
uszm:HashKey rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:HashKey" .
### http://www.usz.ch/rdf/ontology/mapping#SchemaTableName
uszm:SchemaTableName rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:SchemaTableName" .
### http://www.usz.ch/rdf/ontology/mapping#SortIndex
uszm:SortIndex rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:SortIndex" .
### http://www.usz.ch/rdf/ontology/mapping#UniqueKey
uszm:UniqueKey rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:UniqueKey" .
### http://www.usz.ch/rdf/ontology/mapping#SourceFormat
uszm:SourceFormat rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:SourceFormat" .
### http://www.usz.ch/rdf/ontology/mapping#CodingSystem
uszm:CodingSystem rdf:type owl:AnnotationProperty ;
rdfs:label "uszm:CodingSystem" .
Semantic Definition of the Procedure by SPHN CSI WG / dataset
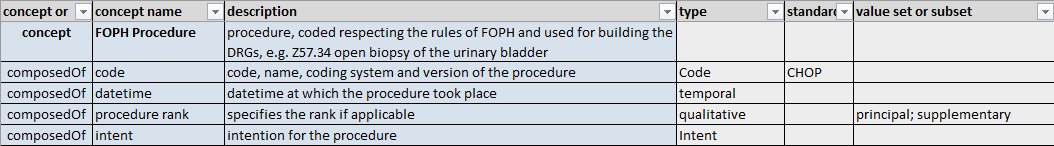
As you can see the first line defines the Concept/Class “FOPH Procedure”, this is defined by the “concept” in the first column The following lines are the Properties/SubClasses the concept is composed of.
Class Mapping
Starting from the underlying class definition
This example shows the definition of the FOPH Procedure as provided by SPHN/DCC:
### https://biomedit.ch/rdf/sphn-ontology/sphn#FOPHProcedure
sphn:FOPHProcedure rdf:type owl:Class ;
rdfs:subClassOf sphn:SPHNConcept ;
rdfs:comment "procedure, coded respecting the rules of FOPH and used for building the DRGs, e.g. Z57.34 open biopsy of the urinary bladder" ;
rdfs:label "FOPH Procedure" .
By convention the class name in the ontology is transformed from “FOPH Procedure” to “FOPHProcedure” by removing all the blanks, substituting special characters with _ (not applicable in this example) and camel casing the words, except for certain abbreviations.
Mapping for the Class
Mapping rules are to be defined on the class level and on the property level. On the class level the information that is mapped are valid for all attributes, like how to generate a unique key for an instance of a FOPH Procedure. This is how the mapping of a class looks like:
# sphn:FophProcedure
#----------------------------------------------------------
usz:FophProcedure-default rdf:type owl:Class ;
rdfs:subClassOf sphn:FophProcedure ;
uszm:HashKey "0"^^xsd:string ;
uszm:Sortindex 3000 ;
uszm:UniqueKey "[UniqueId]"^^xsd:string .
Context
As some classes can be used in different context (i.e. Concept Unit is used for LabResult, DrugAdministrationEvent, HeartRate etc.) the mapping of the class needs to specify for which context it is. As in this example the FOPHProcedure is not used in a different context, the context name that is to be used is ‘default’. This is to be added to the concept name separated by the - sign.
By adding the context to the class name it becomes a different class or sub-class. In order to define that it is linked to the FOPHProcedure the second line predicate “subClassOf” is to be set to the original class name FOPHProcedure.
sphn:FOPHProcedure rdf:type owl:Class ;
rdfs:subClassOf sphn:SPHNConcept ;
Becomes,
usz:FOPHProcedure-default rdf:type owl:Class ;
rdfs:subClassOf sphn:FOPHProcedure ;
uszm:UniqueKey
Every record of an FOPH Procedure becomes and instance of a Node in the RDF File. As a consequence every instance needs to have a unique identifier. This identifier is partially composed automatically by the RDF Extract process and partially by the definition of the class mapping.
<https://biomedit.ch/rdf/sphn-resource/CHE-108.904.325-FophProcedure-221BD46F-E513-4EC6-AE9D-FB31078A3987> or short resource:CHE-108.904.325-FophProcedure-221BD46F-E513-4EC6-AE9D-FB31078A3987
In this Example the resource IRI is composed by:
part A
http://biomedit.ch/rdf/sphn-resource/
or
resource:
implicit Fixed prefix for IRIs of an instances.
part B CHE-108.904.325- implicit This is the ID of the DataProviderInstitute and it is fixed for every university hospital.
part C FophProcedure- implicit Name of the class
part D 221BD46F-E513-4EC6-AE9D-FB31078A3987 explicit
Part D is the value of the predicate UniqueKey. In this case a newID generated by SQL.
The creation of the string for the part 4 is done on the SQL Server side while retrieving the values. As a consequence the mapping rule for the uniqueKey is to be defined as a SQL statement that defines a column in a select statement! The uniqueKey can also be a combination of columns as shown in the example below and it must includes all data type conversions and formatting.
uszm:UniqueKey "[UniqueId]"^^xsd:string .
or composed by different fields like for Frequency
uszm:UniqueKey "[ObservationValue]+'-Unit-'+[ObservationUnitRDF]"^^xsd:string .
uszm:Hashkey (hashing of UniqueKey)
With the introduction of SQL newID as the UniqueKey for each concept the hashing is not needed anymore but it would still be able if needed.
As the unique key of some concepts can be come rather long, the unique key will be hashed in most cases. As this is optional it needs to be define here if the uniqueKey is to be hashed or not. The mapping value can either be 0 (not hashed) or 1 (hashed). As this will be processed as boolean, the load program for the mapping expects this mapping to be of type string (^^xsd:string)
uszm:HashKey "1"^^xsd:string ;
An example of hashed and non hashed key is:
not-hashed: <https://biomedit.ch/rdf/sphn-resource/CHE-108.904.325-Code-CHOP_2020-87.41.99>
uszm:Sortindex
As of RDF schema version 2021.1 the semantic view is sorted and processed by the sorting ConceptName, ClassName, AttributeName and the SortIndex is not used anymore.
Please note also, that the SortIndex is a string value and the data type is optional to be specified if it is a string.
uszm:Sortindex 3000 ;
equal to,
uszm:Sortindex "3000"^^xsd:string ;
Property Mapping
Starting from the underlying property definition, this example shows the definition of the property rank of the FOPH Procedure:
### https://biomedit.ch/rdf/sphn-ontology/sphn#hasFOPHProcedureRank
sphn:hasFOPHProcedureRank rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:SPHNAttributeObject ;
rdfs:domain sphn:FOPHProcedure ;
rdfs:range sphn:FOPHProcedure_rank ;
rdfs:comment "specifies the rank if applicable" ;
rdfs:isDefinedBy <http://snomed.info/id/277367001> ;
rdfs:label "has FOPH procedure rank" .
By convention the property name in the ontology is composed of “has” + ConceptName + PropertyName.
Mapping for the Property
Mapping rules are to be defined on the class level and on the property level. On the property level the information that is mapped are specific to the property. This is how the mapping of a property looks like:
# sphn:FophProcedure-hasFOPHProcedureRank
usz:hasFOPHProcedureRank-FophProcedure rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasFOPHProcedureRank ;
uszm:ColumnName "[ProcedureRank]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[FophProcedure]"^^xsd:string ;
uszm:Filter "[ProcedureRank] is not null"^^xsd:string ;
uszm:Sortindex 110 .
Context
As with classes some properties can be used in different context (i.e. hasDateTime is used for BodyTemperature, BodyHeight, OxygenSaturation etc.) the mapping of the property needs to specify for which context it is. As in this example the hasFOPHProcedureRank is not used in a different context, the context name that is to be used is the name of the concept i.e. ‘FOPHProcedure’. This is to be added to the property name separated by the - sign.
By adding the context’ to the class name it becomes a different property or sub-property. In order to define that it is linked to the superProperty hasFOPHProcedureRank the second line predicate “subPropertyOf” is to be set to the original property name hasFOPHProcedureRank.
sphn:hasFOPHProcedureRank rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:SPHNAttributeObject ;
Becomes->
usz:hasFOPHProcedureRank-FophProcedure rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasFOPHProcedureRank ;
The standard context is “default” for the class name and for properties it is the “{ClassName}”. However, it is important to note that if an extra context for a property needs to be defined an extra mapping for the class needs to be defined with the same context (see example below)!
Following example shows a non-default context for the hasLabResultLabTestCode where there are two different cases that needs to be mapped differently.
Case A (default) is where the LabResultTestCode is expressed as a Loinc code that is linked to an external Taxonomy. In this case it is sufficient to map this directly to the external taxonomy.
Case B (non-default) is where the LabResultTestCode is expressed as another concept “Code”. In this case the mapping has to be adapted to create an IRI that points to a instance of the corresponding Code node.
Class Mapping Property Mapping
Default case
# sphn:LabResult
usz:LabResult-default rdf:type owl:Class ;
rdfs:subClassOf sphn:LabResult ;
uszm:HashKey "0"^^xsd:string ;
uszm:Sortindex 2000 ;
uszm:UniqueKey "[UniqueId]"^^xsd:string .
# sphn:LabResult-hasLabResultLabTestCode-LOINC
usz:hasLabResultLabTestCode-LabResult rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasLabResultLabTestCode ;
uszm:ColumnName "[ObservationCode]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[LabResult]"^^xsd:string ;
uszm:Filter "[CodingSystemVersion] = 'LOINC'"^^xsd:string ;
uszm:CodingSystem "loinc"^^xsd:string ;
uszm:Sortindex 1004 .
Special context
# sphn:LabResult
usz:LabResult-LabResultLabTestCode rdf:type owl:Class ;
rdfs:subClassOf sphn:LabResult ;
uszm:HashKey "0"^^xsd:string ;
uszm:Sortindex 2000 ;
uszm:UniqueKey "[UniqueId]"^^xsd:string .
# sphn:LabResult-hasLabResultLabTestCode-internal
usz:hasLabResultLabTestCode-LabResultLabTestCode rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasLabResultLabTestCode ;
uszm:ColumnName "[CodingSystemVersion]+'-'+[ObservationCode]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[LabResult]"^^xsd:string ;
uszm:Filter "[CodingSystemVersion] = 'internal'"^^xsd:string ;
uszm:CodingSystem "sphn"^^xsd:string ;
uszm:Sortindex 1004 .
uszm:ColumnName
This mapping predicate is defining the column where the value can be retrieved to create to triple-object. This does not necessary correspond to one column in the SQL view. In cases of ObjectProperties which have an IRI as the triple-object it needs to be the same as the UniqueKey of the triple-subbject it links to (see example below).
It might also be necessary to use other SQL functions to manipulate the value of a column i.e. lower(xyz) or columns could be concatenated.
usz:hasLabResultLabTestCode-LabResultLabTestCode rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasLabResultLabTestCode ;
uszm:ColumnName "[CodingSystemVersion]+'-'+[ObservationCode]"^^xsd:string ;
usz:Code-LabResultCode rdf:type owl:Class ;
rdfs:subClassOf sphn:Code ;
uszm:UniqueKey "[CodingSystemVersion]+'-'+[ObservationCode]"^^xsd:string .
uszm:DatabaseName
This value depends on the installation of the RDF extract. At USZ the Database is called “RDFOntology”.
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName
This mapping rule defines on which view the value can be found and usually corresponds to the concept name with the exception of some demographic concepts that are part of view “Patient”. The value should only be the view without the schema name as the schema name is taken from the projectID direclty. This prevents to create project specific mappings.
uszm:SchemaTableName "[FophProcedure]"^^xsd:string ;
uszm:Filter
The filter mapping rule is very important to make sure that a property is only generated if it has a value not equal to NULL or other conditions have to be met. The content of the filter will be added to the SQL WHERE clause when accessing the data. It is important to note that if the filter uses an OR condition it needs to be placed into ( ) or it might create strange results. Please note that every WHERE clause also will have an condition selecting for the correct patient.
SIMPLE:
uszm:Filter "[ProcedureRank] is not null"^^xsd:string ;
COMPLEX:
uszm:Filter "([ActiveIngredient] is not null OR [DrugCodeGTIN] is not null)"^^xsd:string ;
uszm:SoruceFormat
Some of the values are stored in a different format as the ontology defines. By default the value is read in the format as defined by the ontology (i.e. reader.getDouble()) If, however, the column is specified in a different format this needs to be indicated in the mapping with the SourceFormat option. Usually the value would be string but it could also be double or datetime.
uszm:SourceFormat "string"^^xsd:string ;
uszm:CodingSystem
Some properties define different ranges like snomed, atc and sphn:Code in the example below. If no coding system is specified for the mapping all three values would be generated with incorrect values. Therefore the mapping must specify which coding system the mapping corresponds to.
These could be for external taxonomies SNOMED-CT, loinc, atc, chop, icd-10-gm and for SPHN valueset or Code concept it would be sphn. Please note the case sensitivity of this definition!
Description Example for hasSubstanceCode
Ontology allows: snomed, atc, Code
sphn:hasSubstanceCode rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasCode ;
rdfs:domain sphn:Substance ;
rdfs:range [ rdf:type owl:Class ;
owl:unionOf ( <http://snomed.info/id/105590001>
sphn:Code
atc:ATC
)
] ;
rdfs:comment "code, name, coding system and version representing the substance e.g. ATC or SNOMED CT" ;
rdfs:label "has substance code" .
This PropertyMapping is for atc
# sphn:DrugAdministrationEvent-Drug-Substance-SubstanceCode
usz:hasSubstanceCode-DrugAdministrationEventDrugSubstance rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasSubstanceCode ;
uszm:ColumnName "[DrugCodeATC]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[DrugAdministrationEvent]"^^xsd:string ;
uszm:Filter "[DrugCodeATC] is not null"^^xsd:string ;
uszm:CodingSystem "atc"^^xsd:string ;
uszm:Sortindex 1200 .
ATC:
uszm:CodingSystem "atc"^^xsd:string ;
SNOMED:
uszm:CodingSystem "SNOMED-CT"^^xsd:string ;
SPHN Code or SPHN Valueset:
uszm:CodingSystem "sphn"^^xsd:string ;
uszm:Sortindex
As of RDF schema version 2021.1 the semantic view is sorted and processed by the sorting ConceptName, ClassName, AttributeName and the SortIndex is not used anymore.
Please note also, that the SortIndex is a string value and the data type is optional to be specified if it is a string.
uszm:Sortindex 3000 ;
equal to,
uszm:Sortindex "3000"^^xsd:string ;
Linking to DataProvider, Patient and AdministrativeCase
The mapping for DataProviderInstitute, Patient and AdministrativeCase works the same way as for all the other attributes with the exception that it is usually not visible in the SPHN/Project specification Excel and the information for which concepts these links have to be created can be found in the ontology under the corresponding sphn:hasXxxxx definition. For each class listed in the domain a corresponding link mapping is expected.
### https://biomedit.ch/rdf/sphn-ontology/sphn#hasDataProviderInstitute
sphn:hasDataProviderInstitute rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:SPHNAttributeObject ;
rdfs:domain [ rdf:type owl:Class ;
owl:unionOf ( sphn:AdministrativeCase
sphn:AdministrativeGender
...
sphn:FOPHProcedure
...
sphn:TumorSpecimen
sphn:TumorStage
)
] ;
rdfs:range sphn:DataProviderInstitute ;
rdfs:comment "Link to the unique business identification number (UID) of the healthcare institute providing the data" ;
rdfs:label "has data provider institute" .
DataProviderInstitute
The data provider is a bit special in the sense that the value for the data provider is not taken from any view but is initialized at the beginning of the extraction process of the table Map.DataProvider. As a consequence value for the uszm:ColumnName is not relevant. It is suggested to set it to a valid column name, usually [ResearchPatientId] but it could be anything else.
In most of the cases uszm:Filter can be omitted and it only needs to be specified for cases where the concept could not be present and no corresponding node will be generated.
# sphn:FophProcedure-hasDataProviderInstitute
usz:hasDataProviderInstitute-FophProcedure rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasDataProviderInstitute ;
uszm:ColumnName "[ResearchPatientId]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[FophProcedure]"^^xsd:string ;
uszm:Filter "[ProcedureCode] is not null"^^xsd:string ;
uszm:Sortindex 999980 .
SubjectPseudoIdentifier
The mapping for the hasSubjectPseudoIdentifier can usually copied from another concept and replace the Context and the SchemaTableName with the corresponding class.
In most of the cases uszm:Filter can be omitted and it only needs to be specified for cases where the concept could not be present and no corresponding node will be generated.
# sphn:FophProcedure-hasSubjectPseudoIdentifier
usz:hasSubjectPseudoIdentifier-FophProcedure rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasSubjectPseudoIdentifier ;
uszm:ColumnName "[ResearchPatientId]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[FophProcedure]"^^xsd:string ;
uszm:Filter "[ProcedureCode] is not null"^^xsd:string ;
uszm:Sortindex 999990 .
AdministrativeCase
The mapping for the hasAdministratvieCase can usually copied from another concept and replace the Context and the SchemaTableName with the corresponding class.
In most of the cases uszm:Filter can be omitted and it only needs to be specified for cases where the concept could not be present and no corresponding node will be generated.
# sphn:FophProcedure-hasAdministrativeCase
usz:hasAdministrativeCase-FophProcedure rdf:type owl:ObjectProperty ;
rdfs:subPropertyOf sphn:hasAdministrativeCase ;
uszm:ColumnName "[ResearchCaseId]"^^xsd:string ;
uszm:DatabaseName "[RDFOntology]"^^xsd:string ;
uszm:SchemaTableName "[FophProcedure]"^^xsd:string ;
uszm:Filter "[ProcedureCode] is not null"^^xsd:string ;
uszm:Sortindex 999999 .
Load RDF Ontology and Mapping (2021.1)
After installing the C# console program, i.e. compiling from the source, the following directory structure needs to be defined:
RDFData/config
RDFData/logs
RDFData/data
RDFData/scripts
Also the config file needs to be defined in the config directory for the program to run:
# Ontology Load config file
# Project ID according to DeIdMaster.Project.ProjectID
-Project={project_id}
# Run mode, options extract, load
-Mode=Load
# JSON, turtle (RDF *.ttl), possibly later others
-FileFormat=turtle
# data or ontology
-ContentType=ontologyMapping
# timeout for database command execution
-Timeout=500
# input file name, usually the ontology file to load.
-InputFile={pathToMappingFile}\{project_id_}ontology_mapping.ttl
-LogFilePath={pathToLogFiles}\Logs\{project_id}\
# trace level 0 .. 3
# level 0 reports FATAL, ERROR messages
# level 1 reports additionally Warning messages
# level 2 reports additionally Info messages
# level 3 reports additionally Debug messages
-TraceLevel=2
-sphnontologyversion=2021.1
-projectontologyversion=2021.1
-rdfschemaversion=2021.1
Finally before the mapping can be loaded the file {projectId}_ontology_mapping.ttl needs to be generated with the script as documented under Workspace Setup above and copied into the directory as specified in the InputFile path in the config file.
As the ontology/mapping load is a console application you need to open PowerShell and input the following command:
.\DataExtractor.exe -ConfigFile='RDFData\config\ontology_load.config'
6. RDF Extraction into a turtle file
Before the extract can be started the following configuration needs to be set in the database (for the first time only):
INSERT INTO [RDFOntology].[RDF].[Project] ([ProjectID],[ProjectName],[DefaultProject],[RDFSchemaVersion],[ConformsToSPHNVersion],[ConformsToProjectVersion]) VALUES ('{ProjectID}','Sepsis',0,'2021.1','2021.1','2021.8')
For every concept a project likes to receive data for needs to be setup in the table ProjectClass as shown in the following example. The last column IsActive can be used to suppress some columns for certain extracts, especially valuable for testing.
INSERT INTO [RDF].[ProjectClass] ([ProjectID],[ConceptName],[RDFSchemaVersion],[IsActive]) VALUES ('{ProjectID}','HeartRate','2021.1',1)
INSERT INTO [RDF].[Prefix] ([PrefixID] ,[URI] ,[Default] ,[ProjectId] ,[RDFSchemaVersion] ,[isVersionedURI]) VALUES ('{ProjectID}', 'http://biomedit.ch/rdf/{ProjectID}/ontology/', 0, '{ProjectID}' '2020.1', NULL)
Extracting the RDF turtle files is done with the same program as the ontology mapping load and if the program is installed under step 5. Load RDF Ontology and Mapping only the config file for the extract needs to be setup.
# RDF extract config file
# For Project {ProjectID}
-Project={ProjectID}
-Mode=extract
-FileFormat=turtle
-ContentType=data
-rdfschemaversion=2021.1
-FileDirectory={pathToRDFFiles}\RDFData\Data\
-LogFilePath={pathToLogFiles}\Logs\{project_id}\
-BatchSize=4885
-Limit=4885
-FileNamePattern=CHE_108_904_325_|pseudoId|_|project|.ttl
-ByPatient=true
-TraceLevel=2
# The program marks which patient were already extracted. in order to export only the missing patients, the following entry must be set to false or all patients are extracted again.
-ExportAllPatients=true
-Timeout=1000
-sphnontologyversion=2021.1
-projectontologyversion=2021.2
To run the extract program only the config file needs to be specified as all the rest is the same as with running the ongology mapping load.
.\DataExtractor.exe -ConfigFile='RDFData\config\RDF_extract_{project_id}.config'
Now the RDF files should be available in the output directory as specified in the config file.
7. RDF Validation
The validation of the RDF turtle files is checked using the sphn QC framework which can be found here: SPHN Quality Control Framework
However, due to the sizes of some RDF files this is difficult to achieve and each file needs to be validated individually which requires to start the QC pipeline for each file and takes a lot of time to process. As a consequence only random RDF files were checked. This situation will be improved in the future by using the SPHN Connector that automates this process for all RDF files.