Other terminologies
There exist other external resources (e.g., EMDN, ICD-O-3) recommended in the SPHN Dataset for coding values in SPHN besides the ones introduced in the previous sections. These resources are not necessarily provided in the RDF format by the DCC as a single .ttl or .owl file for use.
For these resources, the DCC recommends to provide the codes in the RDF data files with the Code concept with the following attributes:
the identifier
the name
the coding system and version (following conventions defined here)
For example, if an EMDN code A01010199 is provided, the following data instance must be generated by the data provider:
resource:Code-EMDN-A01010199 a sphn:Code ;
sphn:hasIdentifier "A01010199"^^xsd:string ;
sphn:hasName "HYPODERMIC NEEDLES - OTHER"^^xsd:string ;
sphn:hasCodingSystemAndVersion "EMDN-1.1"^^xsd:string .
Find below a (non-exhaustive) list of such external resources with an introduction and possibly information about their use in data science.
EMDN
Introduction to the classification
The European Medical Device Nomenclature (EMDN) is a medical device classification that provides information on the types of medical devices, opposed to product identifier systems with very fine-grained distinction on the level of manufacturer and version or model of a medical device. EMDN is based on the Classificazione Nazionale Dispositivi medici (CND), the Italian nomenclature for medical devices and has been established to support the European database on medical devices (EUDAMED). At the beginning of May 2021, the first version of EMDN was released and manufacturers will be asked to use the EMDN for the registration of medical devices in EUDAMED. An EMDN code will be assigned to each Unique Device Identifier – Device Identifier (UDI-DI) in EUDAMED and the combination of UDI-DI and the associated EMDN code therefore link a product and a type identifier.
The hierarchy of EMDN is divided into seven levels of classification (see example in Figure 1):
Level 1 indicates the category of medical device, referred with a letter
Level 2 indicates the group of medical devices, referred with two numbers
Level 3 to Level 7 indicate the type of medical device, referred with a series of numbers
Figure 1. Visual representation of EMDN codes and their hierarchy.
Category W (in vitro diagnostic medical devices) is of particular interest for research in laboratory diagnostics, as it comprises types of laboratory analyzers, test kits and accessories for in vitro diagnostic devices and tools.
EMDN is used as one of the recommended standard for the SPHN concept:
Lab Analyzer (https://www.biomedit.ch/rdf/sphn-ontology/sphn#LabAnalyzer).
Information for use in data science
The progressive spread and increasing use of LOINC-codes, for example, for clinical observations and laboratory tests, is an enormous benefit for multi-center projects as it increases comparability and interoperability of clinical data. Despite their advantages LOINC often provide no or rather high-level classification on the method employed to generate a laboratory result. Such information, including the type of the medical device(s) used, however, is of major interest for clinical staff and researchers.
Example 1:
EMDN code W02020201 is assigned to coagulometers, and the codes W0202020101 and W0202020102 one level below allow to distinguish semi-automated and automated methods.
Example 2:
EMDN code W0101060101 is assigned to glucose test strips. The hierarchical nature of the EMDN additionally reveals that it is blood test strips (as opposed to, e.g., urine test strips) for rapid and point of care testing.
A type categorization system like EMDN (but also others, e.g., GMDN) can be helpful where different, but similar models of medical devices (e.g. of a model series) are used for analysis in different hospitals, i.e. in cases where a distinction on the level of model adds no further benefit.
GUDID
Introduction to the classification
The Global Unique Device Identification Database (GUDID) contains key device identification information submitted to the FDA about medical devices that have Unique Device Identifiers (UDI). The FDA is establishing the unique device identification system to adequately identify devices sold in the U.S. - from manufacturing through distribution to patient use.
Source: https://accessgudid.nlm.nih.gov/
GUDID is used as one of the recommended standard for the SPHN concept:
Lab Analyzer (https://www.biomedit.ch/rdf/sphn-ontology/sphn#LabAnalyzer).
Information for use in data science
With hospitals and data providers becoming more and more interconnected, e.g., in national or international multi-center projects, data interoperability is of growing importance. The use of LOINC-codes covers clinical observations and laboratory tests, making clinical data collected across different hospitals or laboratories more comparable. LOINCs, however, are often missing information on the method employed to generate a laboratory result, providing no or rather high-level information only. The type of method, analyzer device, or reagent (specified for example by GMDN or EMDN) or even the exact model of a medical device(s) used is of major interest for clinical staff and researchers. When specifying data used to generate reference values for laboratory tests, for example, it is crucial to ensure compatibility of data sources, as therapeutic decisions may rely on them. Specification of devices is unfortunately often still inconsistent between hospitals and laboratories or languages. The use of unique device identifiers (UDIs), e.g., from the Global Unique Device Identifier Database (GUDID) or the upcoming EUDAMED database will be highly beneficial to assign a medical device to a laboratory test without ambiguities.
Example 1
Modern laboratory analyzers are often modular, for example, the Cobas 8000 from Roche featuring several modules for different applications (clinical chemistry and immunoassays) with different rates of throughput. Specifying “Cobas 8000” as medical device used for data collection is therefore not sufficiently granular. Likewise, a human specialist can easily identify designations like “Cobas 8000 c 502”, “Cobas c‑502”, or “c 502 Cobas 8000” as identical, while an automated data processor cannot. A unique identifier (04015630928354) can overcome this issue and in addition link to associated information like manufacturer, catalog number or a device description.
Example 2
Names of laboratory analyses used internally in clinical data warehouses often lack specificity in terms of reagent, test kit or potential supplementary reagents used or required. The assessment of levels of alanine aminotransferase (ALAT), for example, can be done from different sample sources and according to different standards (e.g. with or without addition of pyridoxal phosphate in the reaction). While the former can be specified by the corresponding LOINC (e.g. 1742-6 “serum or plasma” vs. 76625-3 “blood”), the latter can be assigned by the use of a unique identifier for the test kit (04015630920532 “ALAT with pyridoxal phosphate activation” vs. 04015630913862 “ALAT without pyridoxal phosphate activation”).
The combination of product identifiers and type categorization systems like EMDN or GMDN will help to provide information on medical devices with different degrees of granularity, suitable for various purposes.
ICD-O-3
Introduction to the classification
The International Classification of Diseases for Oncology – 3rd Edition (ICD-O-3) is used for coding the body site (topography) and the histology (morphology) of neoplasms. The information is usually obtained from a pathology report and ICD-O-3 is used in cancer registries around the world. ICD-O-3 is provided by the World Health Organization (WHO) and has been updated twice by the International Association of Cancer Registries (IACR) on behalf of WHO (ICD-O-3 1st revision from 2013 and ICD-O-3 2nd revision from 2019).
ICD-O-3 is a multi-axial classification with codes for topography, morphology, behavior and grading of neoplasm (see Figure 1). The topography hierarchy uses mainly the ICD-10 classification of malignant neoplasm. The morphology non-hierarchical axis provides codes composed of histology and behavior. Histologic grading (differentiation) is a separate code.
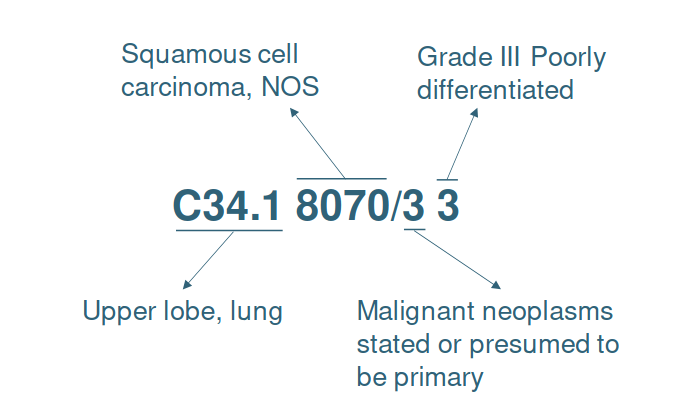
Figure 1. Example of the ICD-O-3 code C34.1 8070/3 3 meaning “Poorly differentiated squamous cell carcinoma, upper lobe of lung”.
ICD-O-3 is used as the recommended standard for the SPHN concept:
ICD-O Diagnosis (https://www.biomedit.ch/rdf/sphn-ontology/sphn#ICD-ODiagnosis).
Versions of ICD-O-3
In Switzerland, ICD-O-3 is used since 2005; the ICD-O-3 1st revision (ICD-O-3.1) was used from 2013 to 2019 and the ICD-O-3 2nd revision (ICD-O-3.2) was used from 2020 onwards.