Omics concepts
This set of concepts is created to represent different types of omics data. The concepts cover clinical use cases from oncology, paediatric care, and pathogen surveillance. You can also check out the Bridging Clinical and Genomic Knowledge: An Extension of the SPHN RDF Schema for Seamless Integration and FAIRification of Omics Data paper, which also describes the concept design and modeling choices.
Concept design
The concepts allow to describe metadata on the omics process, and, in combination with other
concepts, the outcome of the genomics workflow. Also bulk transcriptomics and, to a lesser
extent, omics research in general can be represented. Each step in the omics workflow is a process
concept that is composed of essential metadata about that process. The three top-level concepts for
representing the omics workflow are Sample Processing, Assay, and Data Processing. The
Sample Processing and Data Processing concepts are processes that are executed on some input to
generate some output. The Sample Processing concept can have zero or more input Sample and zero
or more output Sample. Similarly, the Data Processing concept can have zero or more input
Data File and zero or more output Data File.The Sample Processing concept is composed of
zero or more input and output Sample concepts, while the Data Processing concept is composed
of zero or more input and output Data File concepts. The Assay concept, also a process, can
have zero or more input Sample and zero or more output Data File which indicates that the
Assay is a process that transforms an input Sample to an output Data File. As part of the
concept design, we also provided a way to express multi-step processes via the hasPredecessor
property. For example, a process B can link to process A that occurred before itself via the
hasPredecessor property. This style of representation is applicable for Sample Processing,
Data Processing, and in some cases, the Assay concepts.
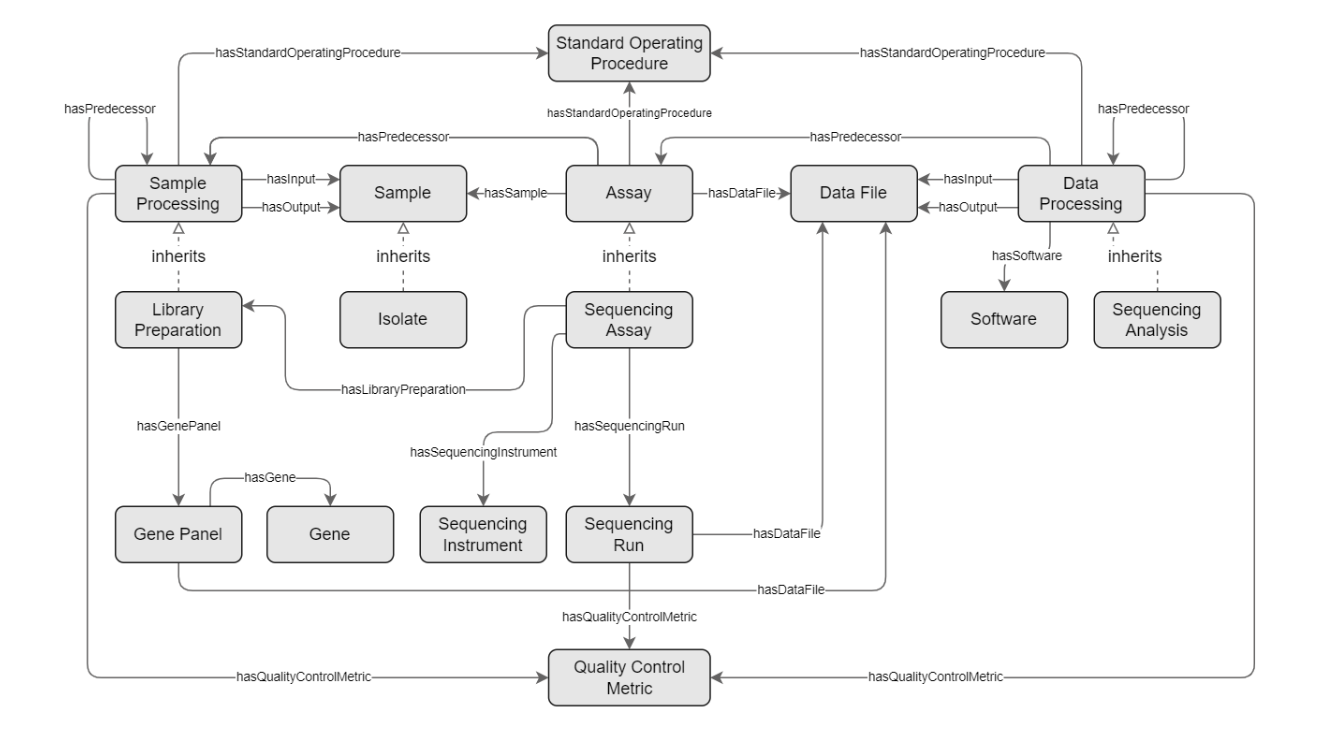
Figure 1. An overview of the (gen)omics concepts
Examples of data delivery
In this section each of the concepts are explained in more detail and examples will be provided.
Assay
Assay metadata is essential when sharing (experimental) results: for providing context, ensuring data quality, enabling data integration, and facilitating collaboration and reproducibility in research and clinical settings. An Assay takes a sample and produces data about that sample. For different types of omics research, different types of assays will be relevant, each with their own defining attributes. The Assay concept can be used as-is, or inherited by more specific types of assay.
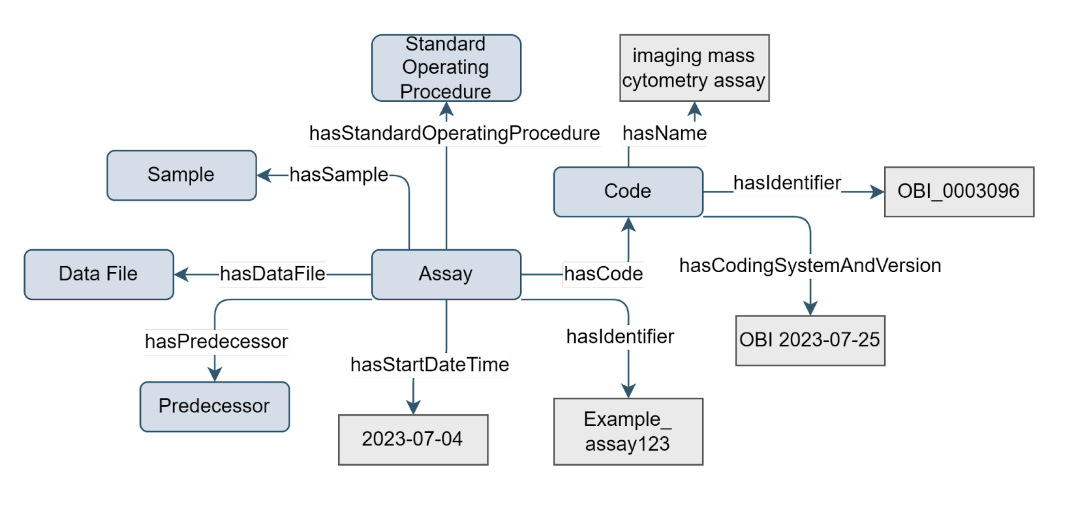
Figure 2. Example of the Assay concept
Guidelines for data delivery
The Assay concept has a
hasCodeproperty which can have a value that is a descendant ofOBI:0000070 | assay |or otherWhen multiple runs are executed for the same assay, as is the case for Whole Genome Sequencing, the start datetime for this concept will be equal to the run datetime of the Run concept that was first executed
All the properties are optional, except for the
hasCodeproperty
Sequencing Assay
Central to the genomics workflow is the sequencing assay. The Sequencing Assay concept is composed of essential metadata, representing the sequencer (via Sequencing Instrument), library preparation (via Library Preparation), intended read length and depth, and zero or more runs (via Sequencing Run). The Sequencing Assay concept is a type of Assay.
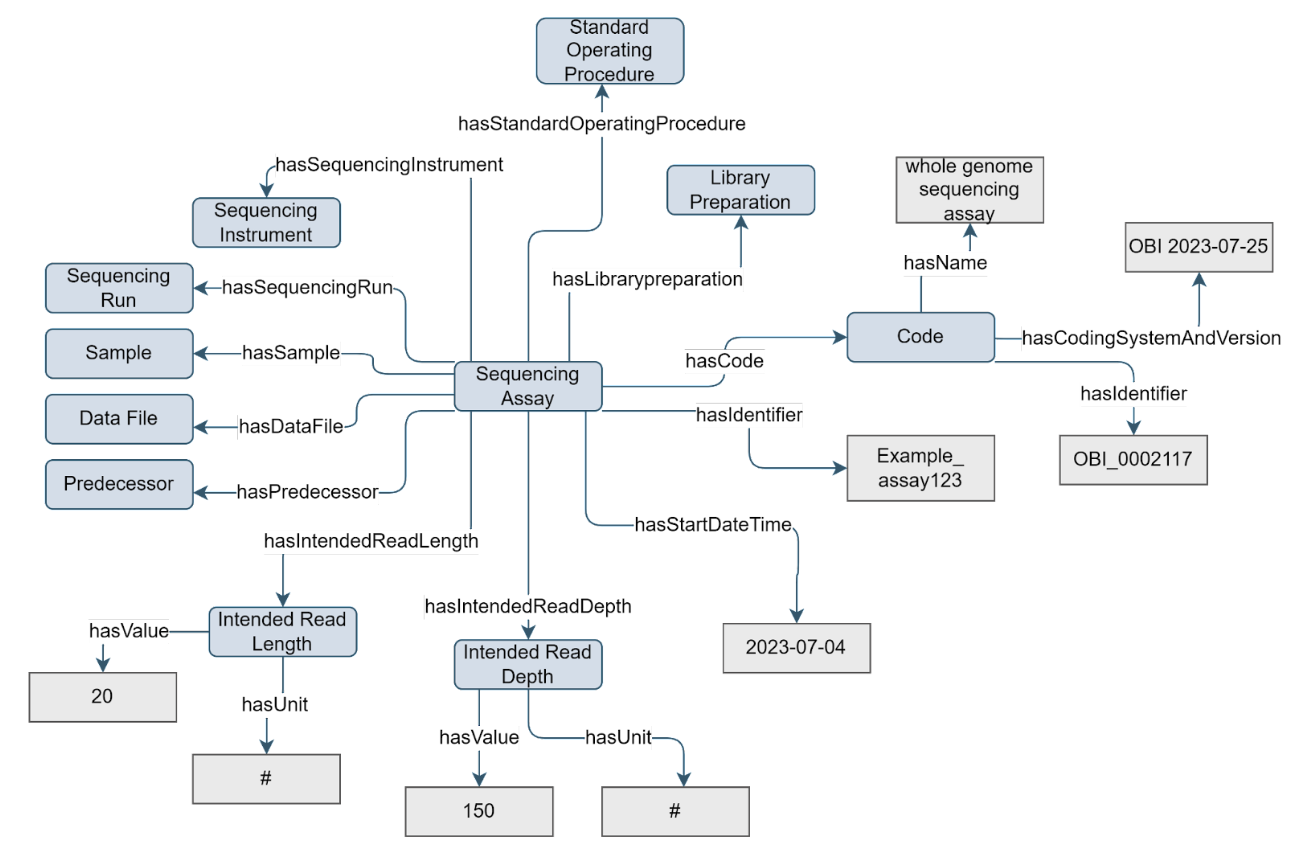
Figure 3. Example of the Sequencing Assay concept
Guidelines for data delivery
A Sequencing Assay may produce multiple Data Files, either different files from a single run or from multiple runs. It is possible to define run-specific information using Sequencing Run, or leave this information out. If a Data File is produced by a Sequencing Run, it follows that it is also related to the linked Sequencing Assay
When multiple runs are executed for the same Sequencing Assay, the start datetime for this concept will be equal to the run datetime of the run that was first executed The Sequencing Assay concept has a hasCode property which can have a value that is a descendant of
OBI:0000070 | assay |or other
Sequencing Run
The Sequencing Run concept represents the actual execution of the assay, and holds information that may vary per run, such as read count, average insert size, average read length, and quality control metrics (represented via the Quality Control Metric concept).
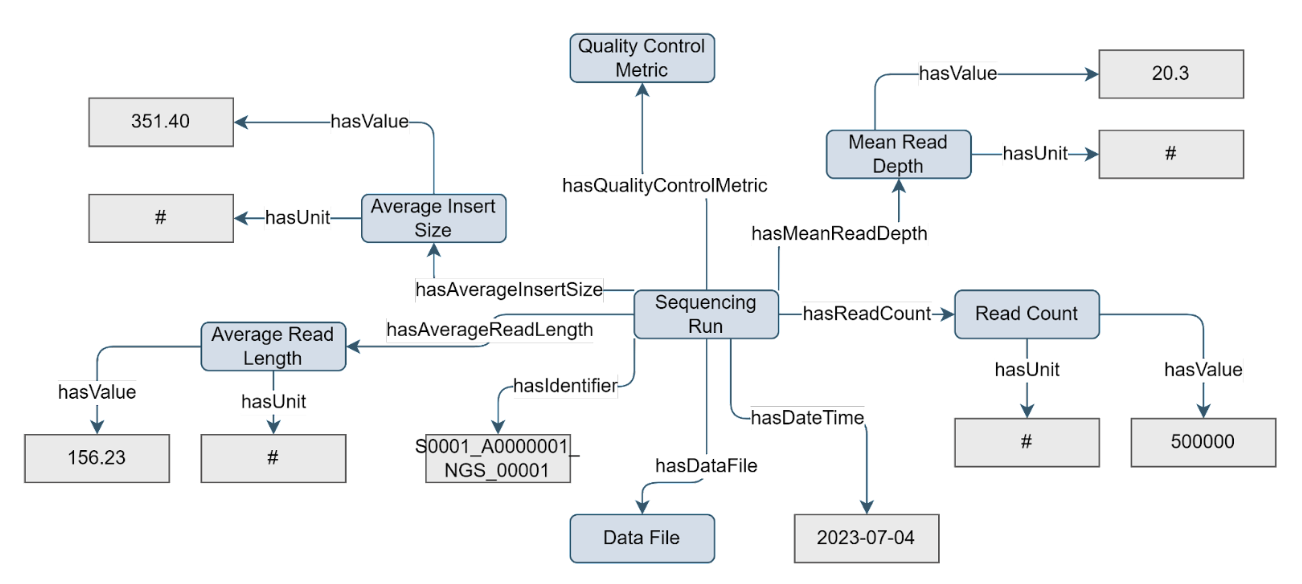
Figure 4. Example of the Sequencing Run concept
Guidelines for data delivery
At least one Data File and Quality Control Metric must be specified. As a result the cardinality for
hasDataFileandhasQualityControlMetricis1:n
Sequencing Instrument
The instrument that was used to conduct a sequencing assay is essential information to understand and evaluate the experimental context and data generation process. Sequencing data may be generated using a range of instruments. Different instruments may vary in sensitivity, accuracy, or precision, and recording the instrument used allows researchers to assess data quality and identify potential sources of variability. Knowing the instrument that was used to produce a dataset enables researchers to assess compatibility and potential biases when performing cross-platform comparisons. The Sequencing Instrument concept contains information about the instrument that was used to conduct a sequencing assay.
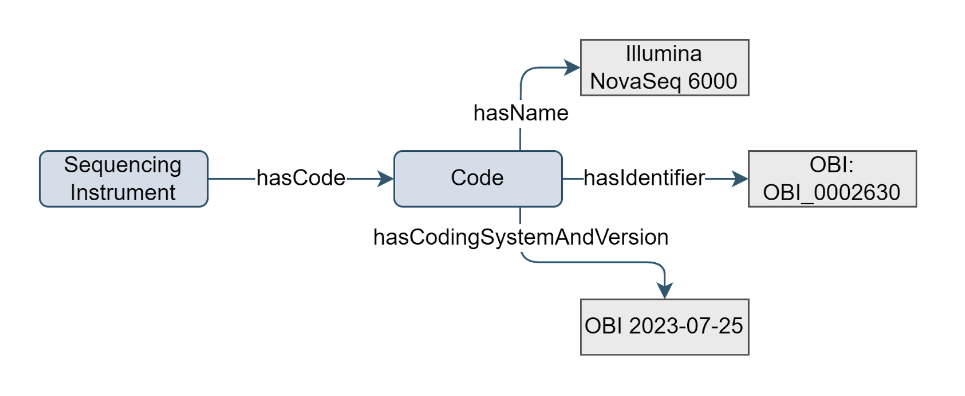
Figure 5. Example of the Sequencing Instrument concept
Guidelines for data delivery
Sequencing Instrument concept has a
hasCodeproperty which can have a value that is a descendant of:OBI:0400103 | DNA sequencer |,EFO:0003739 | sequencer |, or other
Data Processing
An essential part of scientific disciplines is processing the data produced by assays to retrieve an analysis result. Especially in data-intensive domains such as omics, data processing makes up a significant part of the experiment. Usually, individual processing and analysis steps are chained together into a (bioinformatics) pipeline. As part of data processing, data may be transformed from one format or structure to another, or may be subjected to computing to produce aggregates and other analysis results. To evaluate and reproduce these results, metadata on the data processing steps, such as the software/script that was used, is captured in the Data Processing concept.
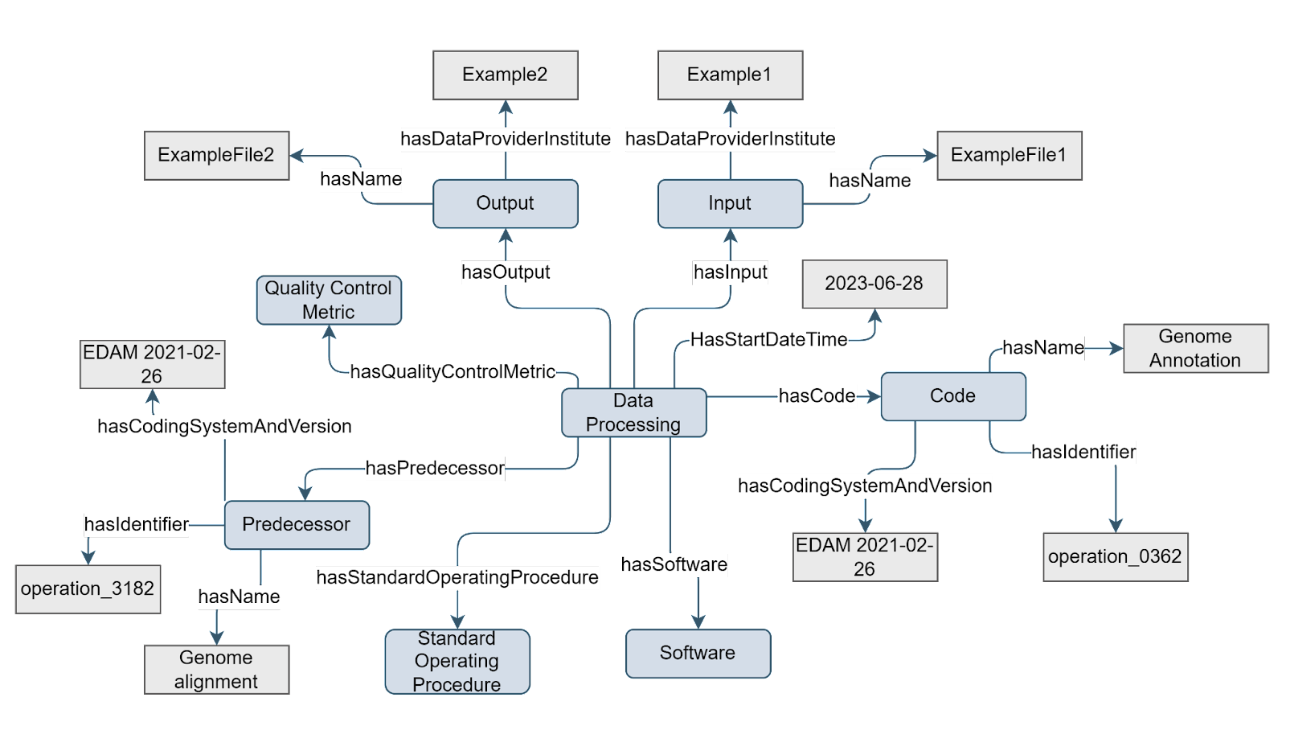
Figure 6. Example of the Data Processing concept
Guidelines for data delivery
The Data Processing concept has a
hasCodeproperty which can have a value that is a descendant ofEDAM:operation_0004 | Operation |,OBI:0200000 | data transformation |, or otherAll the properties are optional, except for the
hasCodepropertyThe Data Processing concept can be used for any data processing step for which the used software should be indicated, such as BCL to FASTQ conversion. The Data Processing concept can be used to indicate sub-steps of a broader process when there is a need to provide metadata for individual steps
Usually, a data processing step has at least one input file. However, there are cases where intermediate files between steps are not known or important. Therefore, the cardinality of
hasInputis0:n
Sample Processing
Sample processing is an essential part of the (omics) experimental workflow. It comprises all processes that manipulate a sample before it can be analysed, such as dissociating tumor cells or culturing. Some sample processing steps are characteristic for a particular omics type, such as library preparation, while others are more general, such as culturing.
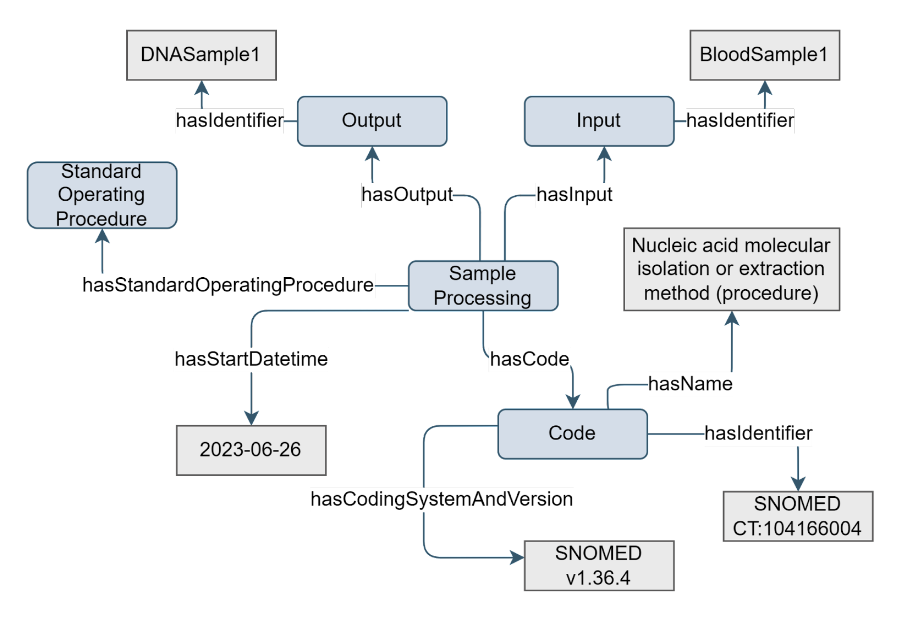
Figure 7. Example of the Sample Processing concept
Guidelines for data delivery
All properties are optional
As with all experimental processes, Sample Processing steps may be chained in sequence, and may consist of individual steps that provide additional metadata. For instance, it can be part of an Assay concept to provide essential metadata on the sample processing that is required for the particular assay type
To indicate the type of sample processing, via hasCode property, descendant terms of
EFO:0002694 | experimental process |,OBI:000011 | planned process |, andSNOMED:71388002 | Procedure (procedure) |can be usedSince Sample Processing has input and output of type Sample, having a composedOf named ‘sample’ is ambiguous. Hence the properties are named
hasInputandhasOutput. Since input and output may not always be relevant or known, for instance in case of intermediate samples between two processing steps, the minimum cardinality of these properties is 0. Note that a sample processing step may have multiple input samples, for instance in the case of a tumor sample and antibody sample, or when multiple input samples are pooled into the same library. However, there can be at most one output sample. When more output samples are output of the same process, for instance when creating slices, repeat this concept for each produced slice sample so that the output is still one sample per sample processhasStartDatetimeis an optional attribute to the Sample Processing concept. As collection datetime is mandatory for Samples, the collection datetime for the output sample is equal to the start datetime of Sample Processing
Sequencing Analysis
NGS sequencing produces raw sequencing data that should be processed and analysed. There are many options to perform this processing and analysis, such as different bioinformatics pipelines and/or scripts (commonly referred to as Software). Metadata about which pipeline and version was used, as well as the used reference genome, are important to compare and evaluate the sequencing results. The Sequencing Analysis concept, a specific type of Data Processing, can be used to store this metadata.
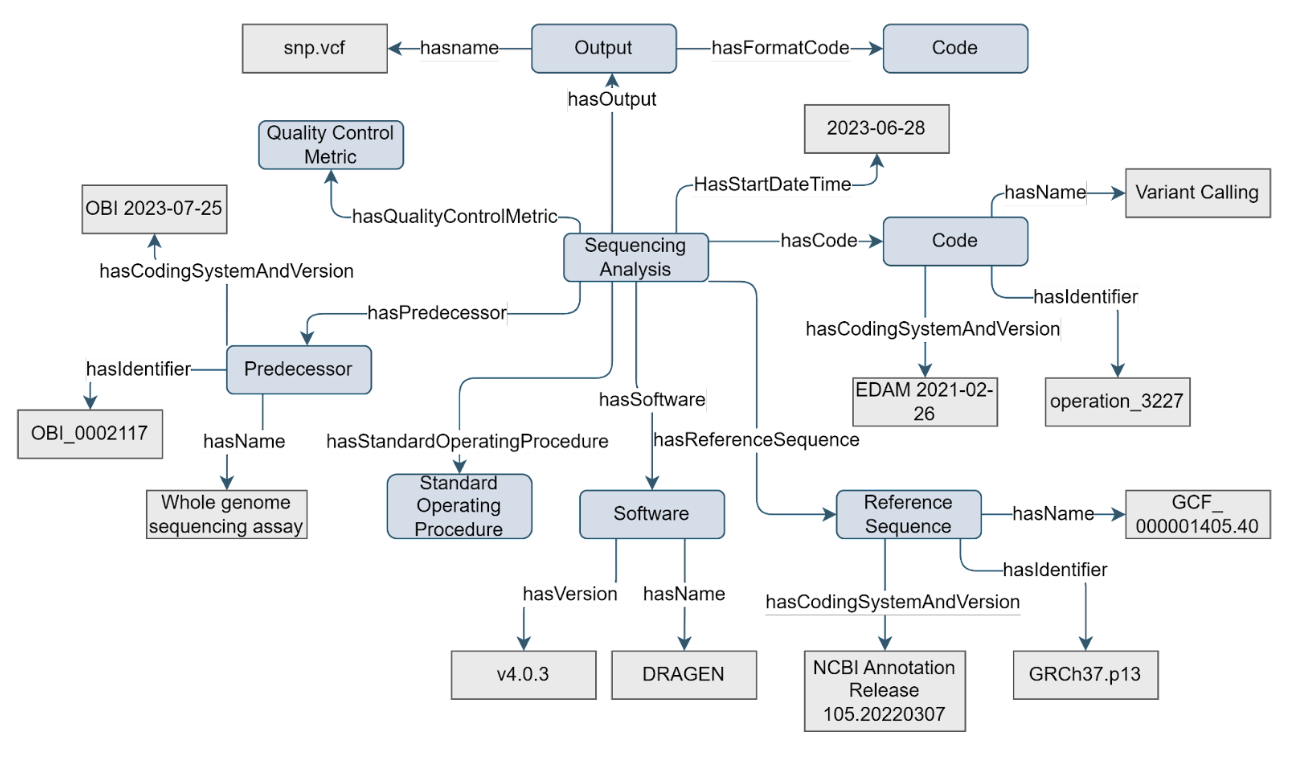
Figure 8. Example of the Sequencing Analysis concept
Guidelines for data delivery
Sequencing Analysis may have many Data Processing parts that are executed one after the other, which can be Sequencing Analysis parts themselves, such as alignment to a reference genome, or more general Data Processing parts, such as data transformation from SAM to BAM files
The Sequencing Analysis concept has a
hasCodeproperty which can have a value that is descendant ofEDAM:operation_2945 | Analysis |or otherThe
hasInput,hasOutput, andhasCodeproperties are mandatory, while everything else is optionalSequencing Analysis is introduced as a special type of Data Processing that always has the aim to analyse data produced by an upstream Sequencing Assay, and uses a reference genome (except in case of de novo assembly). Reference Sequence concept represents the reference genome in case of single organism sequencing, but can be any reference in case of metagenomics sequencing
Standard Operating Procedure
The Standard Operating Procedure (SOP) is a step-by-step description of how experimental procedures should be conducted within an organisation. An SOP is characterised by its name, the textual description, and a version. The purpose of the Standard Operating Procedure concept is to provide the textual description as agreed upon within an organisation, for the protocol followed in an experimental process.
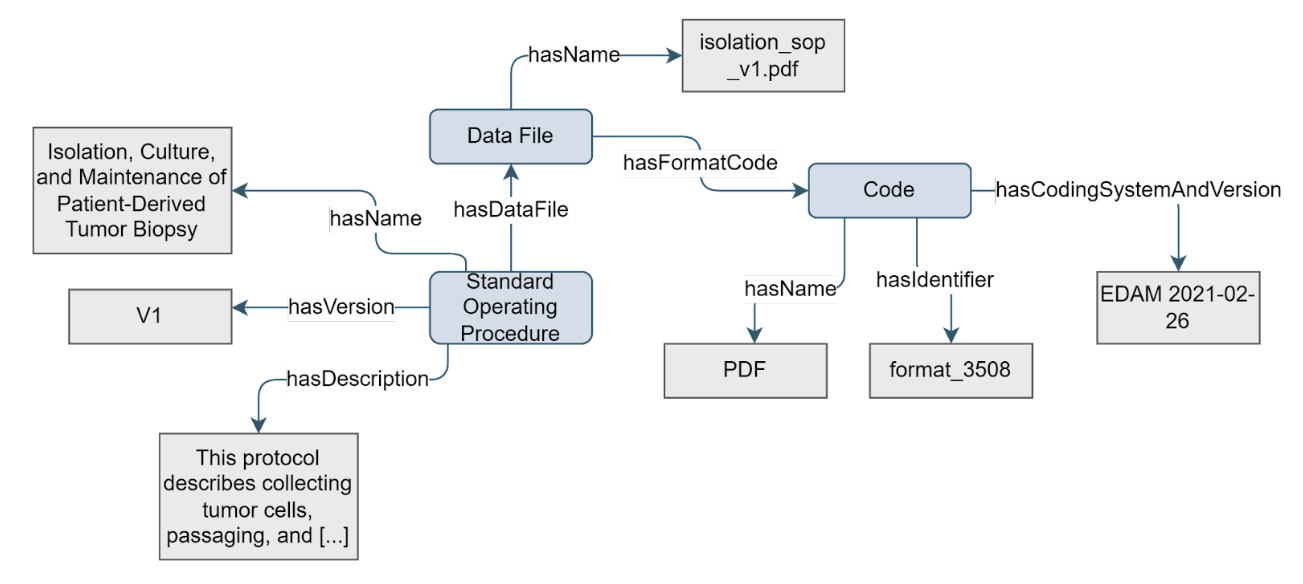
Figure 9. Example of the Standard Operating Procedure concept
Guidelines for data delivery
All properties, except
hasDataFile, have a1:1cardinality. Which means that the SOP must have exactly one name, version and descriptionThe documentation of the experimental steps to follow, or that has been followed, can be indicated as Protocol, or more broadly as Standard Operating Procedure (SOP). SOP has a notion that it is prescribed by an organisation. It also may be broader than experimental protocols
SOPs are available within organisations, but are usually available as documents or text that is not typed or classified, apart from an indication in its name. The experimental process that it prescribes and that it is linked to implicitly types the SOP
Library Preparation
The Library Preparation concept is a type of Sample Processing that is part of a Sequencing Assay. It holds information on the library preparation kit, target enrichment kit, intended insert size, and, in case a gene panel kit is used as target enrichment, information on the gene panel’s focus genes. Any other processing steps that precede an assay’s library preparation may be registered using the Sample Processing concept.
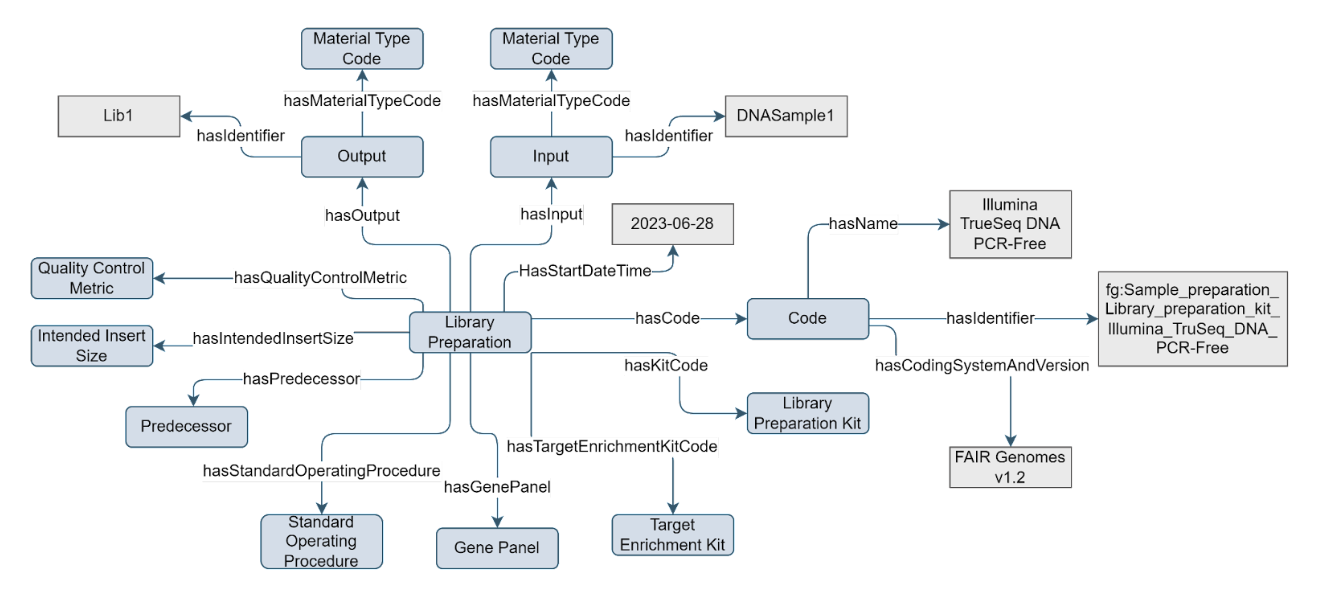
Figure 10. Example of the Library Preparation concept
Guidelines for data delivery
The Library Preparation concept has a
hasCodeproperty which can have a value that is a descendant ofOBI:0000711 | library preparation |, or otherLibrary Preparation can be considered an integral part of performing an NGS sequencing experiment. However, since the configuration/parameters of these experimental processes can vary separately, and also because they can be executed at different facilities at different times by different people, while also having their independent quality control metrics, these are represented as separate concepts that are part of the Sequencing Assay. Note that the linked Sequencing Assay and its Sequencing Instrument are tightly bound to this concept, because these influence the possible choices for the kits used for library preparation and constrains the intended read length and insert size
Note that as with other (experimental) processes, individual sub-steps can be provided, for instance for DNA extraction or amplification
All properties have a cardinality of either
0:1or0:n. This means all properties are optional.
Quality Control Metric
Quality control metrics are used to express the quality of a product or process. They help identify defects, errors, or deviations from established standards. By capturing these metrics in a Quality Control Metric concept, users can ensure that the data meets the quality criteria.
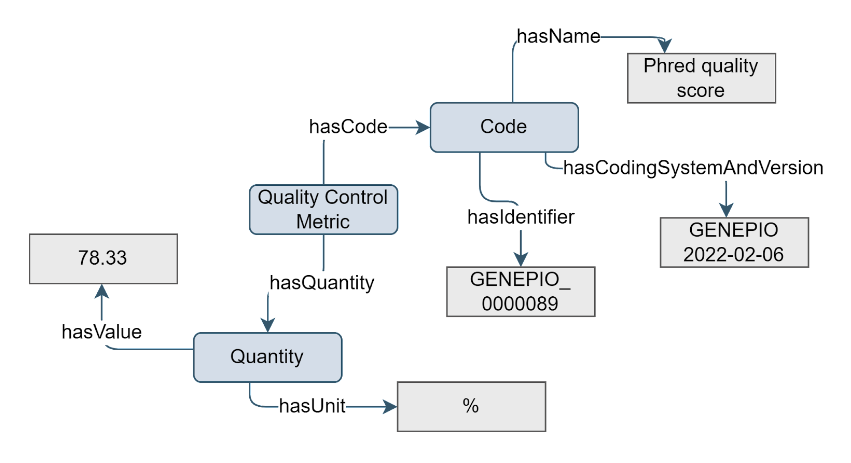
Figure 11. Example of the Quality Control Metric concept
Guidelines for data delivery
Some of the concepts, like Sequencing Run, require a Quality Control Metric. Note that although this information can be mandatory, it is not necessarily guaranteed that data quality will meet or exceed a specific standard; it still needs to be evaluated by the data user, and not all users hold on to the same data quality standards
One or multiple *Quality Control Metric*s can be assigned to a single concept (where applicable)
Isolate
The Isolate concept captures information about specific isolates and their characterization. The Isolate concept is a type of Sample concept.
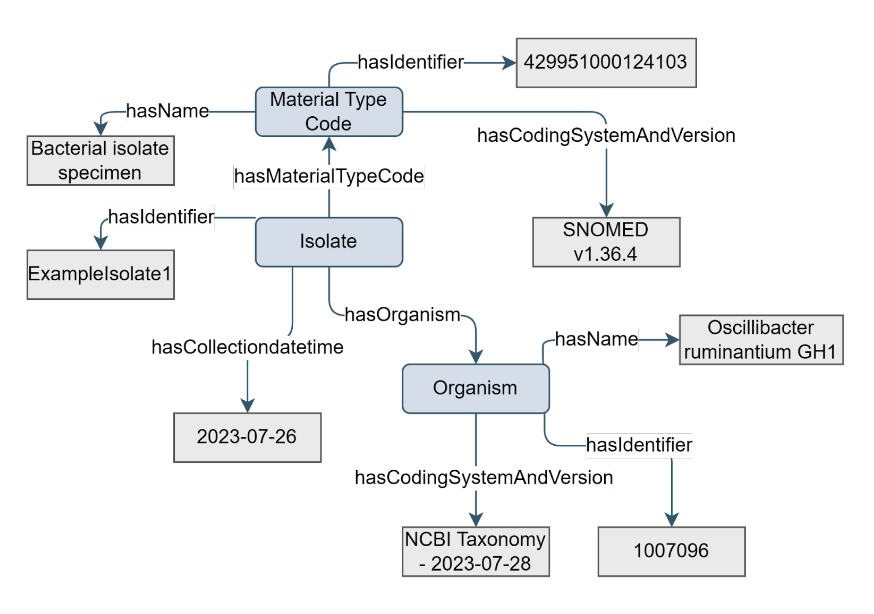
Figure 12. Example of the Isolate concept
Guidelines for data delivery
In contrast to the Sample concept, the Isolate concept is always defined by a species and strain of the pathogen/microbe that is isolated and not of the host
Gene Panel
Gene panels are used for targeted screening in both clinical and research applications. When a gene panel is used for target enrichment as part of Library Preparation, information on the gene panel and its focus genes are required to interpret downstream results. Therefore, the Gene Panel concept can be used to add metadata on the focus genes of the panel to the Library Preparation concept, which is part of a Sequencing Assay.
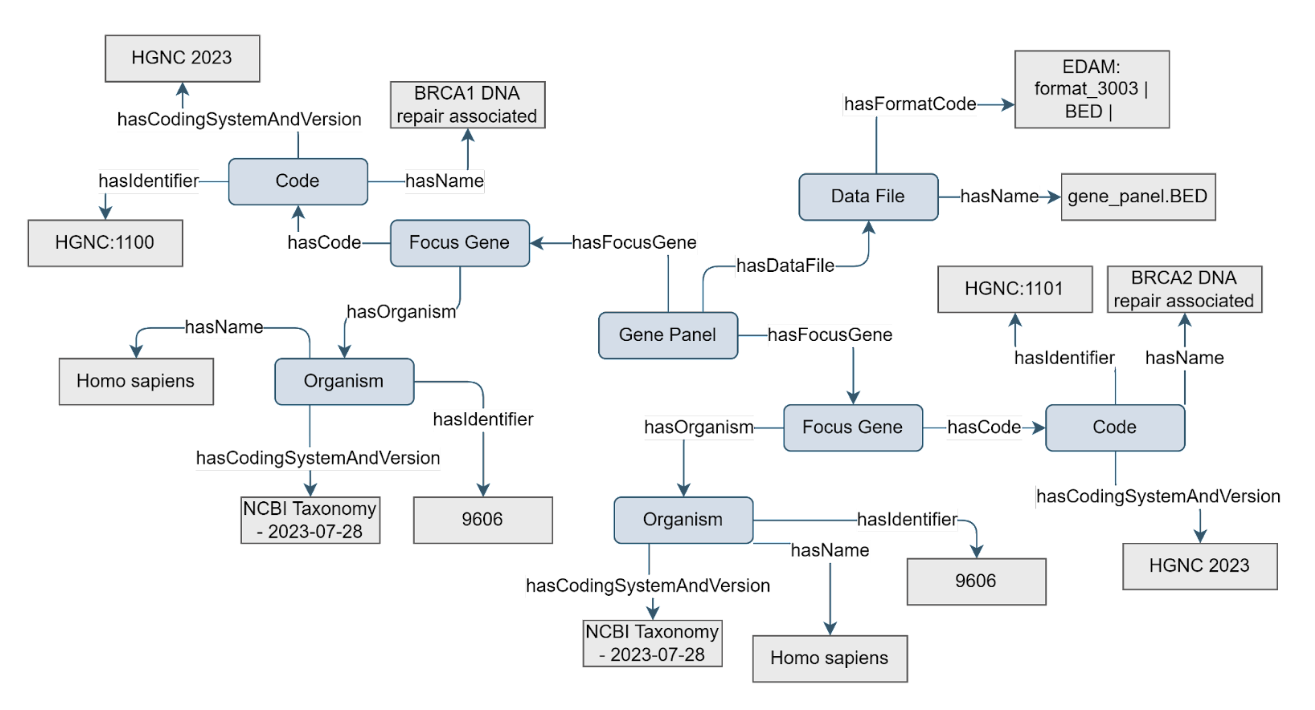
Figure 13. Example of the Gene Panel concept
Guidelines for data delivery
The Gene Panel concept must have at least 1 associated focus gene via the hasFocusGene property
Example of semantic inheritance
For different types of omics research, different types of assays will be relevant, each with their unique set of properties. Therefore, the Assay concept exists. One example of model extension is with a Mass Spectrometry Assay.
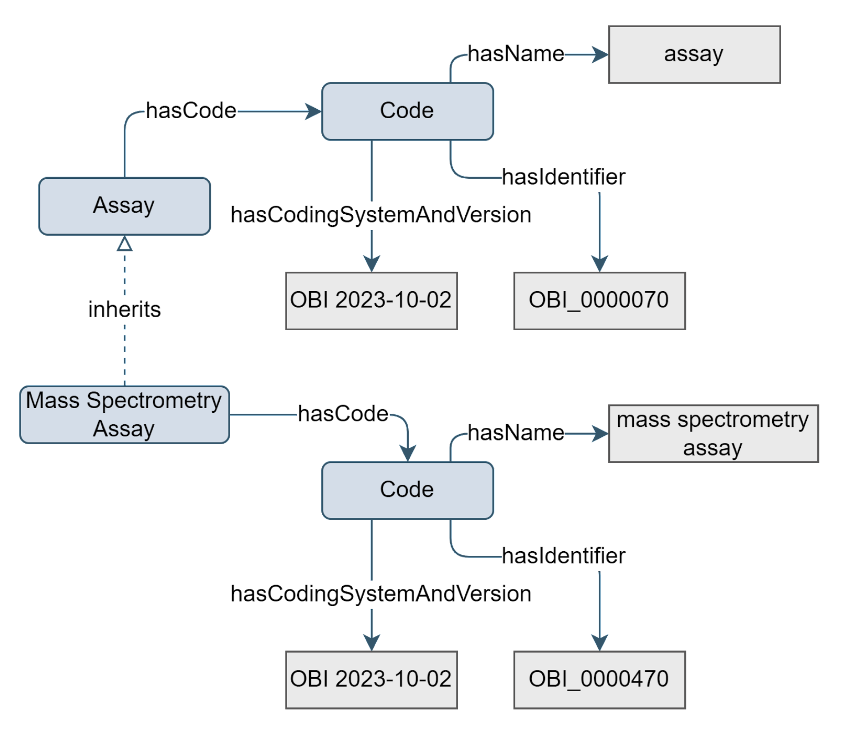
Figure 14. Example of the Assay concept inheritance
Representation of genetic variants
The set of concepts encompassing genomic variants provides the necessary foundational elements for representing variants in a concise and machine-readable format. The chosen representation draws inspiration from GA4GH Phenopackets and GA4GH VRS, aligning with the logical framework of the widely adopted HGVS variant description nomenclature.
Concept design
The design of the concepts for the representation of genomics variants follows the pattern introduced by the VRSATILE framework (a set of conventions extending GA4GH VRS) where descriptors function as central concepts providing all metadata of a specific value concept.
In accordance with this design, the Variant Descriptor concept enables the description of various genetic variations, ranging from simple single point mutations to more intricate structural variations involving large genomic regions. A Variant Descriptor is linked to a specific variant, represented by the Genetic Variation concept. This concept serves as an umbrella, from which all specific variant types inherit (see Figure 1).
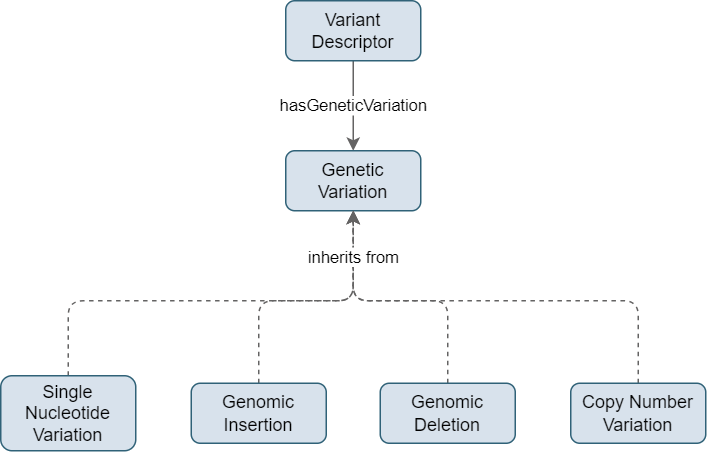
Figure 1. General design of the concepts representing genomic variants.
Variant Descriptor
In addition to capturing essential information about a recorded variant like type, zygosity, or mutation origin, the Variant Descriptor facilitates direct reference to a known variant stored in repositories such as ClinVar or RefSNP, using its code attribute. Genetic variants are commonly described using text strings, varying in complexity according to specific nomenclatures like HGVS, SPDI, or ISCN in the case of structural alterations impacting chromosomes. To accommodate this diversity, the Variant Descriptor can be linked with one or more instances of Variant Notation a flexible concept enabling the representation of a variant description string and its reference notation.
Representing genomic variants using various notations and, consequently, different syntaxes requires users to parse such text strings to effectively query and compare data from diverse sources. For these reasons, variants can be linked to the Variant Descriptor in a machine-readable manner through the Genetic Variation concept.
As a central concept for variant representation, the Variant Descriptor directly connects with the Source System, the Administrative Case, the Data Provider, and the Subject Pseudo-Identifier (see Figure 2).
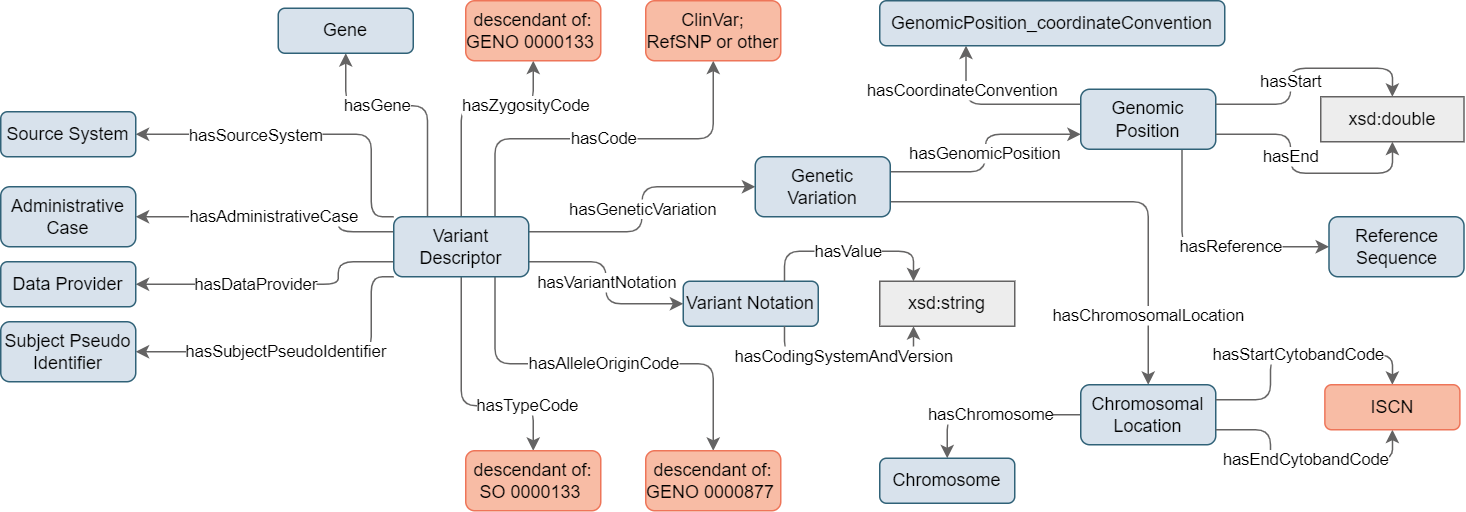
Figure 2. Design of the Variant Description concept showcasing the connection with Variant Notation and Genetic Variation.
Genetic Variation
The concept of Genetic Variation serves as a generic umbrella term encompassing a series of concepts describing genetic variations types. Genetic Variation is comprised of two attributes indicating the position of a variation at the genomic sequence level (Genomic Position) and at a chromosomal resolution (Chromosomal Location) which are inherited by all children concepts.
Specific concepts have been created to cover the most commonly found variants types (see Figure 3):
Single Nucleotide Variation: This concept covers subtle genetic alterations that occur at the level of individual nucleotides within a DNA sequence. These variations involve the replacement of one nucleotide with another, such as adenine (A) being substituted for guanine (G) or cytosine (C) for thymine (T) at a precise location within the sequence.
Genomic Insertion: This concept is integral to a tandem representation of insertions and deletions (indels). It encompasses genetic alterations marked by the addition of one or more nucleotides at a specific location within a DNA sequence. In the current implementation, full support is provided only for insertions of simple contiguous sequences. Complex insertions, such as those involving inverted duplicated copies or the insertion of specific sequences through their reference, are not currently supported. However, it is worth noting that the same information can be captured as the value of the inserted string, albeit with a loss of metadata. Insertions into unknown loci are not currently supported.
Genomic Deletion: This concept is integral to a tandem representation of insertions and deletions (indels). It encompasses genetic alterations marked by the deletion of one or more nucleotides at a specific location within a DNA sequence. In the current implementation, full support is provided only for the deletion of simple contiguous sequences. Genomic deletion is specialized in genomic sequences and, therefore, is not well suited to comprehensively describe deletions occurring in exon/exon, intron/exon, or exon/intron junctions, as well as special cases such as mosaic or chimeric scenarios.
Copy Number Variation: This concept addresses genomic changes involving alterations in the number of copies of a particular DNA segment within an individual’s genome. These variations can encompass both deletions and duplications, leading to deviations in the usual copy number of genomic regions. These variants manifest as relative changes in the quantity of entire genomic segments. Examples include the deletion or duplication of entire genes or larger chromosomal regions.
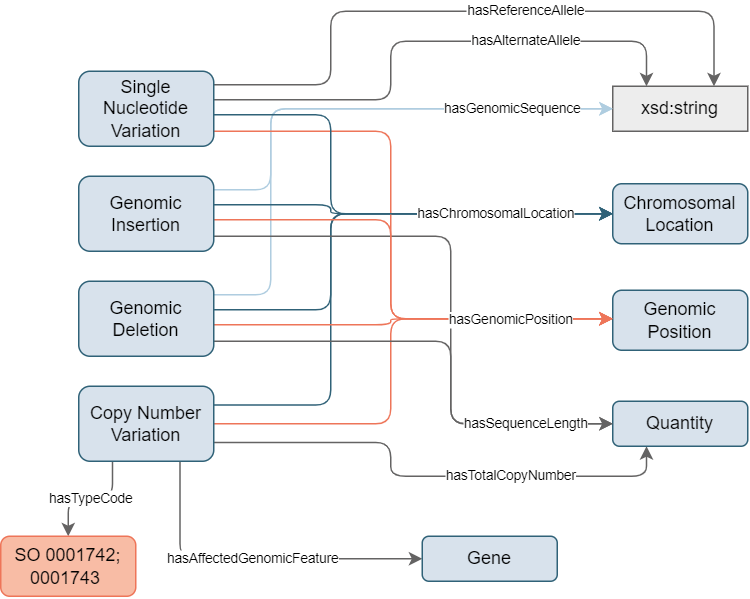
Figure 3. Design of the Variant Description concept showcasing the connection with Variant Notation and Genetic Variation.
Examples for data delivery
In the first example (see Figure 4), a patient underwent genomic analysis targeting genes involved in lung cancer. One of the variants observed pertains to the Epidermal Growth Factor Receptor (EGFR) gene and results in a likely benign mutation of a single nucleotide at a specific locus. To describe this variant, it is necessary to instantiate Variant Descriptor and link this instance with an instance of the Single Nucleotide Variation concept. The Variant Descriptor instance contains comprehensive information about the variant, which, in this case, is of the ‘substitution’ type. Since the sample used for the analysis is derived from a tumor sample, the allele originates from somatic cells (‘somatic allele origin’). Furthermore, it is observed to affect both alleles at a specific locus, indicating ‘homozygous’ zygosity. The allele is documented and registered in public archives like ClinVar, and the accession number is provided to facilitate crosslinking.
The variant’s computable representation is established through the instantiation of the Single Nucleotide Variation concept. This involves specifying the precise locus where the variation occurs (with respect to a reference sequence) and, ultimately, indicating the specific nucleotide that undergoes substitution ‘G>T’.
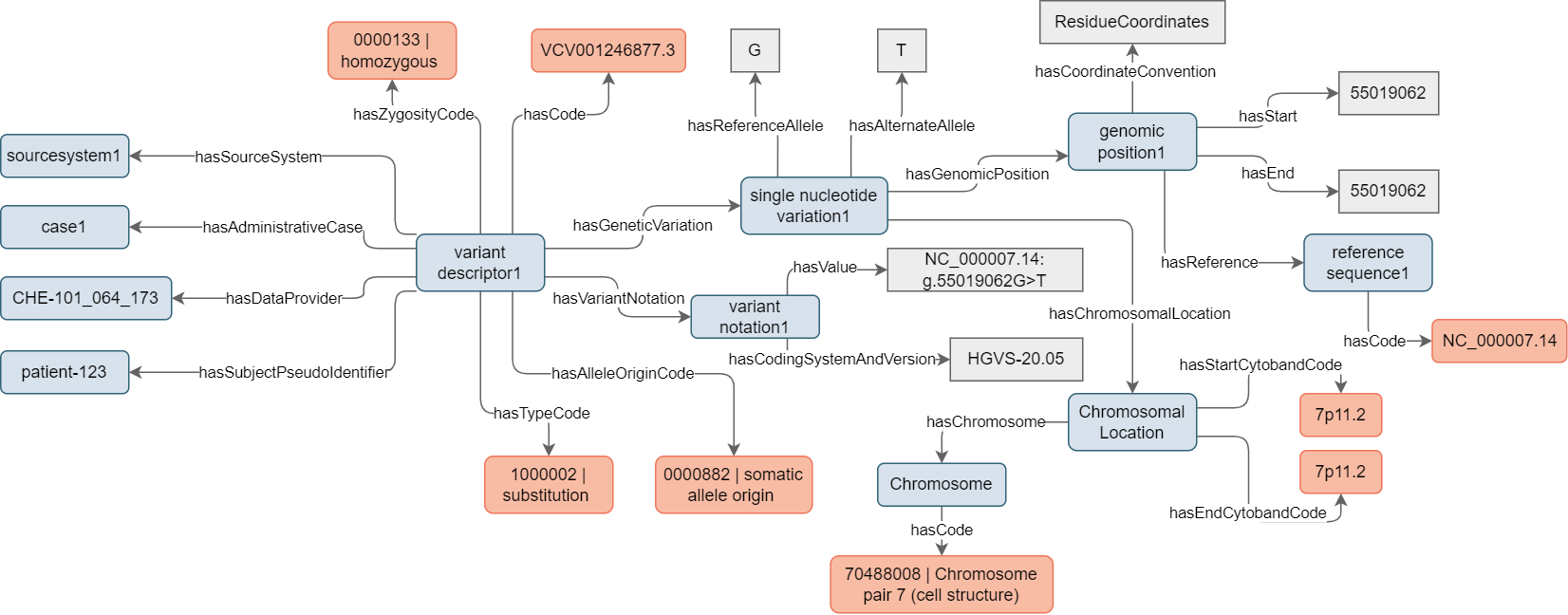
Figure 4. Example of mock instantiation of a Variant Descriptor and Single Nucleotide Variation to describe a SNV discovered in a patient genetic test.
Similar information can be provided by an instance of Variant Notation directly linked to the Variant Descriptor. In the first example, this allows providing a complementary description of the SNV using the widely known HGVS notation. However, in the second example (see Figure 5), Variant Notation can be used to describe genetic variations not covered by a specific concept. In this case, a genetic analysis for a patient highlighted a translocation between chromosome 2 and chromosome 3, with specific breakpoints on the long arm of chromosome 2 at band q13 and on the short arm of chromosome 3 at band p25. Using the ISCN nomenclature, this results in t(2;3)(q13;p25), a common mutation occurring in tumors originating from the thyroid follicular epithelium.
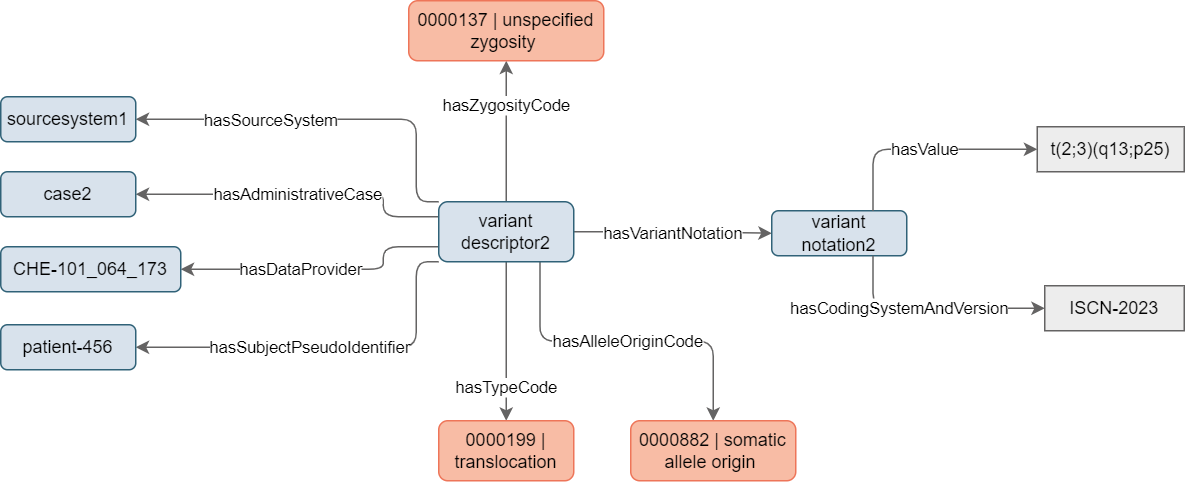
Figure 5. Example of mock instantiation of a Variant Descriptor to describe a translocation discovered in a patient genetic test.
Guidelines for data delivery
Variant Descriptor
The Variant descriptor concept can be used to describe:
A candidate diagnosed variant.
A variant result of a specific molecular test (e.g., sequencing or genotyping).
A candidate variant in specific molecular test (e.g., targeted variant).
When describing a variant, an instance of Variant Descriptor should always be present. This might or not be linked to a more detailed description of the variation using Genetic Variation. The code field allows a variation to be linked to external sources like the ClinGen allele registry, ClinVar, dbSNP, dbVAR, and others.Genetic Variation.
Genetic Variation
The concept of Genetic Variation serves as an overarching framework encompassing various types of genetic variations. Under this umbrella, specific types of genetic variations such as Single Nucleotide Variation, Genomic Insertion, Genomic Deletion, and Copy Number Variation are categorized.
Genetic Variation itself should not be directly instantiated but rather serves as a parent concept, guiding the organization and understanding of specific genetic variations.
Proper usage entails employing the specific child concepts to describe genetic variations,
Genomic positions and chromosomal locations
In the current model, locations or loci are defined as precise positions within a sequence, with specific start and end numerical coordinates, or broad locations represented by chromosomal bands. This information is conveyed through the concepts of Genomic Position and Chromosomal Location, which are core attributes of the Genetic Variation concept. Each specific variant type concept inherits Genetic Position and Chromosomal Location from their common parent concept. The cardinality of these attributes allows a certain degree of flexibility in indicating the position of the variant:
Genetic Position should be instantiated when the exact coordinates of a variation are known.
Chromosomal Location is intended for representing cytogenetic results (e.g., karyotyping).
It is possible to omit positional information altogether when it is unknown.
Chromosomal Location
In the current implementation, Chromosomal Location is intended solely for human chromosomes. For use with different species, both the nomenclature for cytoband representation (currently ISCN) and the standard for describing chromosomes (currently SNOMED CT) should be extended.
For events occurring within a specific cytoband, both the start cytoband and end cytoband must be instantiated with the same value.
For events spanning across multiple chromosomal locations (e.g., large deletions), the interval must represent a contiguous region within the same chromosome. According to GA4GH VRS, the order in which cytoband coordinates are represented is p-terminus → centromere → q-terminus orientation. Consequently, bands on the p-arm are represented in descending numerical order when selecting cytobands for start and end.
Genomic Position
The concept of genomic position allows for the precise representation of coordinates within a continuous reference sequence defined by the Reference Sequence concept. An essential feature of this concept is the ability to choose the preferred coordinate system by instantiating the “Coordinate Convention” attribute, which can be set to either “Residue” or “Inter-residue” coordinate conventions (see Figure 6):
Residue Coordinate Convention: When the Residue coordinate convention is selected, each nucleotide is assigned a specific position along the sequence. This convention, commonly used in systems like HGVS or VCF, provides a straightforward representation of nucleotide positions.
Inter-residue Coordinate Convention: In contrast, the Inter-residue coordinate convention, introduced with the GA4GH VRS, defines positions between nucleotides. This system is particularly useful in scenarios involving deletions or insertions, as it offers a more precise representation of genomic coordinates and reduces ambiguity in genomic data interpretation.
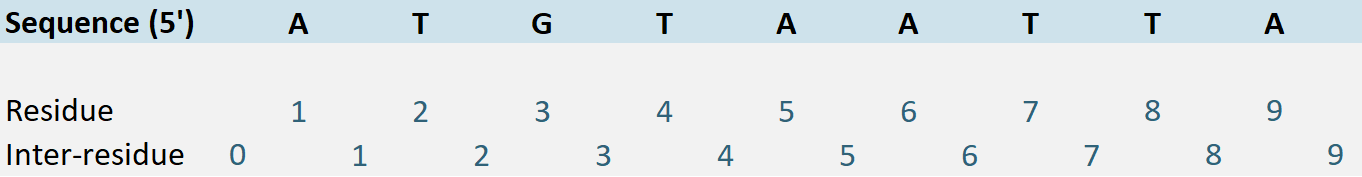
Figure 6. Coordinate conventions in use within Genetic Position.
In addition to that, the following rules applies to concept:
The minimum range for both Start and End attribute is ‘0’.
The End attribute value must be greater o equal to the Start attribute.
As for GA4GH VRS, Genomic Position consider that all locations are with respect to the positive/forward/Watson strand.