Generate a project-specific RDF Schema
Target Audience
This document is intended for project data managers and researchers interested in generating their project-specific RDF Schema. Guidance on how to create a project-specific RDF Schema from a Dataset Template is given.
Introduction
A SPHN project can extend existing (SPHN) concepts and create new concepts (referred to as semantics in the following paragraphs) to fit their needs. Note that under no circumstances a project can modify existing content of the SPHN Dataset. The extension of the semantics for project-specific needs implies that the project must generate its own project-specific RDF Schema. This project-specific RDF Schema will be shared by the project to data providers to get data compliant with their new schema. The project-specific RDF Schema always extend the content (i.e. semantics) defined in the SPHN RDF Schema.
There exists two ways for a project to extend the SPHN semantics and produce their RDF Schema (see Figure 1):
Figure 1: The two options to generate a project-specific RDF Schema.
Option 1: from the SPHN Dataset Template (Excel file with content of the SPHN Dataset) provided by DCC, the project extends the file with the semantics it needs. The project then passes the modified SPHN Dataset Template (which becomes the project-specific Dataset) as input to the SPHN Schema Forge to produce the project-specific RDF Schema automatically
Option 2: the project defines its semantics and directly edits the SPHN RDF Schema with any editor of its choice, compliant with Semantic Web technologies (e.g. Protégé), to produce the project-specific RDF Schema manually.
Procedure to update the semantics
In both options, the procedure for updating the semantics is the same. Figure 2 shows the process that must be followed when using and modifying the content of the SPHN Dataset (semantics) to fit the project-specific needs. The project can reuse existing SPHN concepts, extend SPHN concepts or create new concepts. When modifying existing concepts or building new concepts, these changes have the possibility to be integrated in the future within the SPHN Dataset. SPHN projects are required to design a new concept or modify an existing concept according to the Guiding principles for concept design.
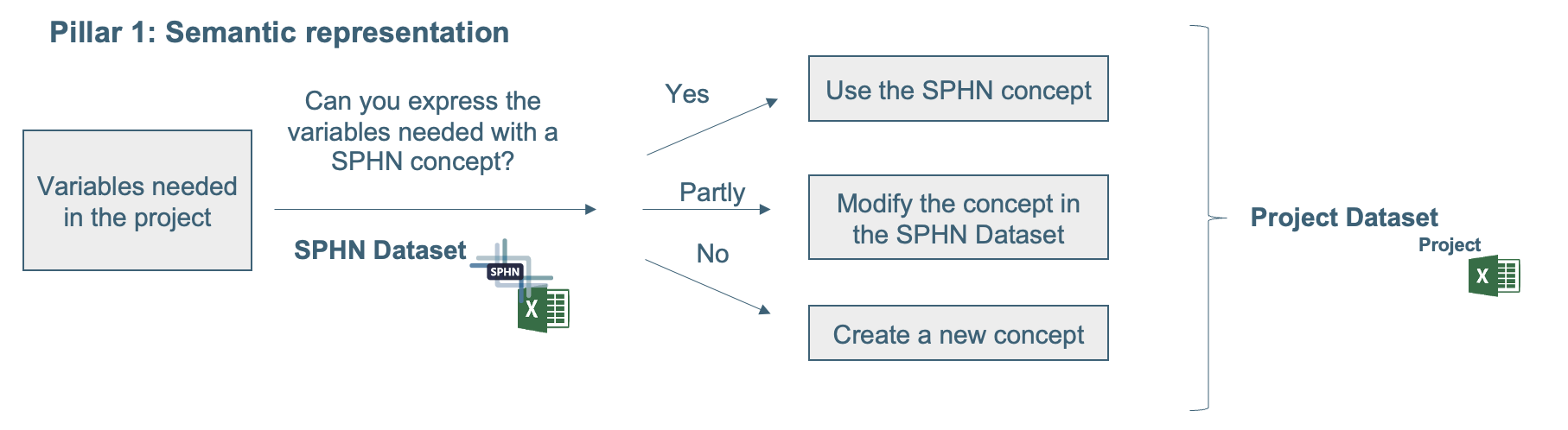
Figure 2: Process on how to use and modify the SPHN Dataset for the project-specific needs.
The extension or modification of existing SPHN concepts can result in additional composedOfs, an alternative semantic standard that needs to be added, or it can be a required extension of an existing value set. There are various reasons calling for extensions, e.g. implementation of a new standard in the applicable jurisdiction, change in availablity of biomedical data, new needs of research projects, or expanded medical knowledge.
Note
There exist four SPHN concepts that have a special meaning in the processing:
Subject Pseudo Identifier, Data Provider, Administrative Case and Source System
Any extension or modification of these concepts might result in invalid pipelines.
Please inform DCC if you want to modify these concepts.
It may happen that you find the concept in the SPHN Dataset for the data you need,
but a piece of information is missing. For example, you need to know the location where a specific measurement is taken,
e.g. Body Temperature Measurement. However, this is not defined in the SPHN concept of Body Temperature Measurement.
In this case you can extend the SPHN concept with the additional composedOf in
your project-specific Dataset.
Note
If you create an extension for your project, please submit a corresponding change request to the DCC via dcc@sib.swiss. A change request template is available on https://git.dcc.sib.swiss/sphn-semantic-framework/sphn-schema/-/tree/master/templates. The extension might be relevant to other projects. The DCC can coordinate an extension to the SPHN Dataset if needed.
Example of semantic extension
description |
type |
||
|---|---|---|---|
concept |
Body Temperature Measurement |
body temperature of the individual |
|
composedOf |
result |
measured temperature |
Body Temperature |
composedOf |
start datetime |
start datetime of the measurement |
temporal |
composedOf |
end datetime |
end datetime of the measurement |
temporal |
composedOf |
body site |
body site of the measurement |
Body Site |
composedOf |
method code |
method code used to measure the temperature |
Code |
composedOf |
medical device |
medical device used to measure the temperature |
Medical Device |
composedOf |
performer |
performer of the measurement |
Performer |
composedOf |
location |
location of the measurement |
Location |
For the example above, one possible next step would be to define your value set or subset for the new composedOf.
Location, as an SPHN concept, already holds a type code property with a value restriction.
In case you are choosing to restrict further the existing value set of Location type code,
in the context of a Body Temperature Measurement to be only in ‘Hospital’, you are allowed to do so.
description |
type |
value set or subset |
||
|---|---|---|---|---|
composedOf |
location |
location of the measurement |
Location |
type code restricted to: 276339004 |Environment (environment)| |
Note
The notation used in the value set is explained in 5. Defining value set or subsets.
The semantics to be integrated in the project must be defined before going to the technical implementation detailed below with the two options to produce in fine the project-specific RDF Schema.
Option 1: Produce an RDF Schema from the SPHN Dataset Template
The Dataset Template is provided as an Excel sheet to be modified by projects to extend the SPHN Dataset according to their needs.
Definitions of terms used in the Dataset (i.e., concept, composedOf)
can be found in the Guideline sheet of the Dataset Template
but also in SPHN Dataset.
Once the Dataset Template Excel file is opened, do the following:
1. Add project’s metadata
Select the sheet Metadata and add the following information below the already filled SPHN metadata line:
prefix: define the prefix that will be used in your project
title: provide a short title about the dataset
description: provide a short description of the content of the dataset
version: the version of the dataset you are building. It should be in the form of
<year>.<number>prior version: if any, provide the previous version of the dataset
copyright: provide information about the copyright of the dataset
license: provide the iri of the license under which the content of the dataset and the schema belong to
canonical_iri: provide the full canonical iri of the dataset that will be created
versioned_iri: provide the versioned iri of the dataset that will be created. It should match the version information provided in
version.
Example
A project called “Genotech” that wants to fill the Dataset Template, starts by providing its metadata:
prefix |
title |
description |
version |
prior version |
copyright |
license |
canonical_iri |
versioned_iri |
|---|---|---|---|---|---|---|---|---|
genotech |
The Genotech project Dataset |
The Dataset of the Genotech project, based on the SPHN Dataset 2024.1 |
2024.1 |
© Copyright 2024, Genotech Institute |
https://www.biomedit.ch/rdf/sphn-schema/genotech# |
Note
The Genotech project builds a Dataset for the first time, therefore the ‘prior version’ field is left empty.
2. Add information about coding system
The DCC provides information about terminologies, standards, vocabularies, and ontologies - henceforth collectively referred to as “coding systems” - that can be used in the SPHN Dataset for representing particular values with codes from the coding systems.
These information are given in the Coding System and Version sheet.
Some of these coding systems are provided in RDF, either by the original provider of the coding system or by the DCC,
while others are not. In case of the latter, one would represent codes from such coding systems as instances of Code concept in the data.
Since 2023.3 release of the SPHN Dataset Template, the Coding System and Version sheet
has been updated to incorporate additional information about coding systems used in SPHN and SPHN projects.
Note
This sheet is also updated in the SPHN Dataset 2024.2 release.
The intention for this change was to:
clarify and differentiate which coding systems are used and/or provided in SPHN and SPHN projects,
facilitate the import of coding systems in RDF by the Dataset2RDF
To that end, a project must update the Coding System and Version sheet to integrate information about supported
and used coding systems in their projects independent of whether or not they are provided in RDF.
If your project mentions a coding system that the SPHN Dataset does not, please add it to the list.
If you are providing a coding system in an RDF version that the DCC is not, please add it to the list and fill
the appropriate columns.
Following are the columns from the Coding System and Version sheet that can be populated:
short name: common abbreviation of the coding system
full name: full name or title of the coding system
coding system and version: short name of the coding system followed by a pattern that represents the way the coding system is versioned by the provider
example: example of an existing version of the coding system (that conforms to the pattern expressed in the ‘coding system and version’ column)
provided in RDF (yes/no): indicate whether the coding system is provided by the project in RDF
downloadable in RDF (yes/no): indicate whether the coding system is downloadable in RDF from any location on the web (typically from the original provider)
provided by: the name of the project (should be the same as the prefix written in 1. Add project’s metadata) to indicate that this coding system is provided/used in the project (i.e. the coding system is not provided/used in the SPHN Dataset and is specifically needed for the project)
prefix: prefix of the coding system, typically corresponds to the ‘short name’ from the ‘short name’ column
root node: indicate the root node that will be used to group all concepts from the coding system in RDF. The coding system may have multiple root nodes, in which case list them all separated by a semi-colon
canonical iri: IRI of the codes taken from the coding system or defined by the project (it can be a
biomedit.ch-based iri if the coding system does not have a web-resolvable IRI for their codes)resource prefix: if applicable, a specific resource node can be created to group all codes (including the root node) under a resource node. For this resource node, a specific prefix must be given
resource iri: if applicable, the IRI for the resource node. The iri must be of the form
https://biomedit.ch/rdf/sphn-resource/.... This column goes hand in hand with ‘resource prefix’ columnversioned iri: the versioned IRI of the coding system. This IRI is used to import the coding system in the RDF schema
Examples
ATC - provided in RDF by DCC
In SPHN, the ATC coding system is actively being used and provided in RDF by the DCC.
ATC codes have dereferencable links which is encoded via the ‘canonical iri’ column.
However, a root node is created in order to group all ATC codes under the same parent.
This root node is defined as ATC and uses the IRI from the ‘resource iri’ column.
Information about ATC is provided as follows:
short name |
full name |
coding system and version |
example |
provided in RDF (yes/no) |
downloadable in RDF (yes/no) |
provided by |
prefix |
root node |
canonical iri |
resource prefix |
resource iri |
versioned iri |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
ATC |
Anatomical Therapeutic Chemical classification |
ATC-[YEAR] |
ATC-2023 |
yes |
no |
SPHN |
atc |
ATC |
sphn-atc |
ORPHA - provided in RDF on the web
ORPHA is a coding system that provides codes that represent rare diseases.
Let’s assume that the Genotech project wants to use the ORPHA and aims to provide the ORPHA codes in RDF. ORPHA is already listed in the SPHN Dataset Template but it is not provided in RDF by the DCC.
During the investigation phase, the Genotech project members discover ORDO (Orphanet Rare Disease Ontology) which represents ORPHA codes in a structured way and compliant with Semantic Web standards. This ORDO ontology fits their needs.
Therefore, the Genotech project would like to use the ORDO ontology and will provide it in RDF.
The Genotech project can then update the line containing ORPHA to add metadata about the coding system as follows:
short name |
full name |
coding system and version |
example |
provided in RDF (yes/no) |
downloadable in RDF (yes/no) |
provided by |
prefix |
root node |
canonical iri |
resource prefix |
resource iri |
versioned iri |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
ORPHA |
Orphanet nomenclature of rare diseases |
ORPHA-[YEAR]-[MONTH] |
ORPHA-2021-07 |
yes |
yes |
GENOTECH |
orpha |
ORPHA |
sphn-orpha |
https://www.orphadata.com/data/ontologies/ordo/last_version/ORDO_en_4.3.owl |
Note
The ‘canonical iri’ corresponds to the IRI used for ORPHA codes in the ORDO ontology.
The ‘resource iri’ and ‘resource prefix’ are internal to the Genotech project (and defined in the context of SPHN) in order to group all the content from the ORDO ontology under the same root node ORPHA.
The ‘versioned iri’ follows the way ORDO is versioned; here it corresponds to version 4.2 of the ORDO ontology.
NANDA - provided in RDF by the project
NANDA is an example of a coding system which is neither downloadable in RDF nor provided in RDF by the DCC. The project first needs to “FAIRify” and translate the coding system into RDF as much as possible (see FAIRify external terminologies in RDF) before using and sharing it.
Again, lets assume that the Genotech project wants to use NANDA and decides to provide it in RDF.
The following metadata is encoded in the Coding System and Version sheet:
short name |
full name |
coding system and version |
example |
provided in RDF (yes/no) |
downloadable in RDF (yes/no) |
provided by |
prefix |
root node |
canonical iri |
resource prefix |
resource iri |
versioned iri |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
NANDA |
NANDA Nursing Diagnoses |
NANDA[YEAR-YEAR] |
NANDA-2018-2020 |
yes |
no |
GENOTECH |
nanda |
NANDA |
Note
In this example, the ‘resource prefix’ column and ‘resource iri’ column do not need to be defined because the root node (NANDA)
and all resources of NANDA will share the same namespace since resources from NANDA do not have a properly defined and dereferencable link.
Coding systems’ copyright
It is important to keep in mind that before providing any coding system in RDF and eventually sharing them with data providers and/or other data users, the project has the responsibility to check the applicable terms and regulations for using and sharing the coding system.
Copyright information must be stated in the RDF terminology file generated/used in the context of the project. This is an important step that should not be neglected.
The use and sharing of a coding systems’ file in RDF in the context of a project is the responsibility of the project data manager. In the future, if the DCC integrates that coding system in the SPHN Dataset and provides it in the Terminology Service, then it will be under the responsibility of the DCC.
3. Concept definition
The next step is to go to the Concepts sheet which already contains all concepts and composedOfs defined in SPHN.
A project is allowed to only:
extend an existing SPHN concept with a project-specific composedOf
- create a new project-specific concept
add existing SPHN composedOfs to it (reuse of existing composedOfs)
add project-specific composedOfs to it
A project is not allowed to:
edit any existing line of an SPHN concept and its related SPHN composedOfs
delete any line of an SPHN concept
2.1 Add a new project-specific concept
A project can decide to add a new concept to their dataset. A concept is an idea or notion that represents, in the context of SPHN, clinical-, health- and genomic-related elements. A concept here can be compared to the notion of “class” in other fields.
To add a new concept, insert a new line at the end of the SPHN Dataset Template and fill the following columns:
release version: version of the dataset when the concept is created or last modified
IRI: the versioned IRI ending with the concept name in UpperCase convention (it must point to the latest version of the project-specific schema)
active status:
yesornoare the allowed values. You must selectyesfor an active conceptconcept reference: provide the name of the concept being created with the following notation:
<prefix>:<Concept Name>concept or concept compositions or inherited: indicate by selecting one of the three options if the row corresponds to a concept, a composedOf or if it is a composedOf that is inherited from another concept. In this case, this cell must be filled with
conceptgeneral concept name: provide the general name of the concept which will be used in the RDF schema
general description: provide the general description of the concept which will be used in the RDF schema
contextualized concept name: provide the contextualized description of the concept in the particular context it is used (this information is not carried on the RDF schema)
contextualized concept description: provide the contextualized description of the concept explaining its meaning in the particular context it is used (this information is not carried on the RDF schema)
parent: provide the general concept name of the parent with the following notation:
<prefix>:<ConceptName>
Note
Unlike in the
Concept referencecolumn, theParentis written in an UpperCase convention without space!The root concept in SPHN is called
SPHNConcept. It contains all concepts defined in SPHN. All SPHN concepts are children of theSPHNConceptbut some concepts can be children of another SPHN concept, which becomes their parent concept. The child concept must then have a more specific meaning than the parent concept. Similarly, all project concepts should be children of the project root concept, which is defined as:<prefix>:<PREFIX>Concept. Same as in the SPHN Dataset, multiple levels of hierarchies can be created. Therefore, a project concept can be the child of another project (or sphn) concept if it has a more specific meaning.
type: Only to be filled if there exist a parent concept which is not the general
<prefix>:<PREFIX>Conceptmeaning binding: if available, a meaning binding of the concept to an coding system can be provided to further anchor the meaning of the concept defined in the project with external resources
additional information: text can be added to provide details to the reader of the dataset (this information is not carried on the RDF schema)
cardinality for the concept to Administrative Case: provide the cardinality of the concept with respect to the Administrative Case by keeping in mind the following: how should the instance of this concept be expected to be linked to the Administrative Case?
cardinality for the concept to Data Provider: provide the cardinality of the concept with respect to the Data Provider by keeping in mind the following: how should the instance of this concept be expected to be linked to the Data Provider?
cardinality for the concept to Subject Pseudo Identifier: provide the cardinality of the concept with respect to the Subject Pseudo Identifier by keeping in mind the following: how should the instance of this concept be expected to be linked to the Subject Pseudo Identifier?.
cardinality for the concept to Source System: provide the cardinality of the concept with respect to the Source System by keeping in mind the following: how should the instance of this concept be expected to be liked to Source System? does it make sense?
Note
You can highlight a concept by making the line bold for an easier reading.
2.2 Add a new project-specific composedOf
Once a concept is created, a project can add project-specific composedOfs to this concept. A project can also add a new project-specific composedOf to an existing SPHN concept. In the latter, two options are possible: either add a new line below the SPHN Concept and its SPHN composedOfs. write in this new line the project-specific composedOf. OR add a new line at the end of the Dataset Template. Copy the line with of the SPHN Concept of interest. Add below the new copied line, the project-specific composedOf for this SPHN Concept.
ComposedOf can be considered as metadata of a concept (i.e., specific information about the concept) and can be compared to properties or attributes of a concept.
The following columns in the Dataset Template can (and should whenever possible) be filled:
release version: version of the dataset when the composedOf is created or last modified
IRI the versioned iri ending with the composedOf name in lowerCase convention
active status: select
yesfor a new composedOf added to a conceptconcept reference: provide the name of the concept this composedOf belongs to with the following notation:
<prefix>:<Concept Name>concept or concept compositions or inherited: indicate by selecting in the list either
composedOforinheritedif the composedOf is inherited from another conceptgeneral concept name: provide the general name of the composedOf which will be used in the RDF schema with the following notation:
<prefix>:<composedOf>general description: provide the general description of the composedOf which will be used in the RDF schema
contextualized concept name: provide the contextualized description of the composedOf in the particular context it is used (this information is not carried on the RDF schema)
contextualized concept description: provide the contextualized description of the composedOf explaining its meaning in the particular context it is used (this information is not carried on the RDF schema)
parent: provide the general composedOf name of the parent with the following notation:
<prefix>:<composedOf>
Note
It is important to note that unlike in the
Concept reference, theParentis written in a lowerCase convention without space.Their exist two root attributes in SPHN for composedOfs:
SPHNAttributeDatatypefor datatype attribute composedOfs andSPHNAttributeObjectfor object attribute composedOfs. Similarly, parents of project’s composedOfs should be pointing to one of the project root attribute<prefix>:<PREFIX>AttributeDatatypeor<prefix>:<PREFIX>AttributeObjectwhen the composedOf is not a descendant of another one.
type: provide the type of the composedOf (e.g., quantitative, qualitative, Code, any SPHN/project concept)
excluded type descendants: list the concepts, separated with a semi-colon, which are not considered as valid types (must be done for concepts where the type is a parent concept but children are not valid types)
standard: when the type of the composedOf is
Code, a coding system can be referenced to indicate possible values. Indicate the name of that coding system in this columnvalue set or subset: when the type of the composedOf is
Codeorqualitative, a set of values or subset of values can be specified in this column
Note
Indicate a subset by starting with
descendant of:followed by the identifiers/values.Indicate a value set by listing the values and separating them with a semi colon
;.The standard nomenclature to write codes from coding system is:
<coding system name>: <identifier> |<label>|. More details on the possible patterns can be found in 5. Defining value set or subsets.
additional information: text can be added to provide details to the reader of the dataset (this information is not carried on the RDF schema)
cardinality for composedOf: indicate the range of cardinality for the composedOf with respect to the concept for which it is defined by keeping in mind the following: when the concept is instantiated we expect this cardinality to be true.
Examples
Example 1: The Genotech project would like to add the concept of Cost to their project.
The concept Cost is described by a value (an existing composedOf in SPHN) and a currency code (a new project-specific composedOf).
The Dataset Template would be filled as follow:
a new line is created for the concept
Costwith the following information:
release version: 2024.1
IRI: https://www.biomedit.ch/rdf/sphn-schema/genotech/2024/1#Cost
active status: yes
concept reference: genotech:Cost
concept or concept compositions or inherited: concept
general concept name: genotech:Cost
general description: an amount that has to be paid or spent to buy or obtain something
contextualized concept name: genotech:Cost
contextualized concept description: an amount that has to be paid or spent to buy or obtain something
parent: genotech:GENOTECHConcept
meaning binding:
additional information:
cardinality for concept to Administrative Case:
cardinality for the concept to Data Provider:
cardinality for the concept to Subject Pseudo Identifier:
cardinality for the concept to Source System:
Note
This line gives the information about a Concept called Cost
which do not have any link to the Administrative Case,
Data Provider or Subject Pseudo Identifier. It is possible to have a concept X
that is not connected to any of these three concepts, in which case the concept X
must be reused in another concept in a composedOf.
a new line is created below the
Costfor adding the composedOfvalue:
release version: 2024.1
IRI https://www.biomedit.ch/rdf/sphn-schema/sphn/2024/1#hasValue
active status: yes
concept reference: genotech:Cost
concept or concept compositions or inherited: composedOf
general concept name: value
general description: value of the concept
contextualized concept name: value
contextualized concept description: value of the cost paid or spent
parent: SPHNAttributeDatatype
type: quantitative
standard:
value set or subset:
additional information:
cardinality for composedOf: 1:1
The composedOf ‘value’ exists in SPHN, therefore, the project can reuse it.
Note
value is a composedOf used in the context of Cost (i.e., concept reference).
With the cardinality, the project indicates that a Cost must have at least one and only one value connected.
a new line is created below the
valuefor adding the composedOfcurrency code:
release version: 2024.1
IRI https://www.biomedit.ch/rdf/sphn-schema/genotech/2024/1#hasCurrencyCode
active status: yes
concept reference: genotech:Cost
concept or concept compositions or inherited: composedOf
general concept name: genotech:currency code
general description: currency of the concept
contextualized concept name: currency
contextualized concept description: currency of the value paid or spent
parent: genotech:GENOTECHAttributeObject
type: Code
standard: ISO 4217
value set or subset:
additional information:
cardinality for composedOf: 1:1
The composedOf ‘currency code’ has not been defined in SPHN, therefore it is created under the project’s IRI.
However it is of type: Code which is an SPHN concept.
Note
currency code is a composedOf used in the context of Cost (i.e., concept reference) and
is of type Code from the SPHN Dataset. With the cardinality, the project indicates that a
Cost must have at least one and only one currency code connected.
Example 2: SPHN Concept extension with project-specific composedOf
The Genotech project wants to add a cost to an Administrative Case to retain information
about the billing costs of a case.
The project has already created a new concept Cost but now wants
this information to be a part of Administrative Case.
The following line can be added in the Dataset Template either directly below the Administrative Case concept and its SPHN composedOfs or at
the end of the Dataset Template after copying the Administrative Case concept line:
release version: 2024.1
IRI https://www.biomedit.ch/rdf/sphn-schema/genotech/2024/1#hasCost
active status: yes
concept reference: Administrative Case
concept or concept compositions or inherited: composedOf
general concept name: cost
general description: cost of the concept
contextualized concept name: cost
contextualized concept description: cost written in the administrative case
parent: genotech:GENOTECHAttributeObject
type: genotech:Cost
standard:
value set or subset:
additional information:
cardinality for composedOf: 0:1
4. Inheritance of SPHN Concepts
A project can define an SPHN concept being the parent of a project-specific concept. This is called inheritance: the project concept has then a more specific definition of the SPHN parent concept (see more about semantic inheritance as it is defined and used in the SPHN Dataset here: Semantic inheritance).
The rule is that when a project concept inherits from an SPHN concept, it must inherit all the properties of that SPHN concept. In the Dataset Template this means a project concept which has as parent an SPHN concept will list the composedOfs of that SPHN concept under the project concept as “inherited” (text to be selected in column ‘concept or concept composedOf or inherited’) composedOfs. In this case, the project has the possibility to narrow down the set of values allowed for a given inherited composedOf.
Note
Note 1: When a inherited property has for type “Code”, the SPHN Dataset usually “restricts” the coding systems
to be used to X, Y, Z or other. The “or other” in principles enables the
project to use any Terminology (coding system they provide in RDF format, codes used with IRIs)
or Code (coding system not provided in RDF, codes used as Code) without breaking the
SPHN Dataset logic.
Note 2: Terminologies provided in RDF in a project-specific Dataset will be listed in the project-specific RDF Schema under
a project:Terminology class. The project:Terminology class must be a subClass of sphn:Terminology.
The project:Terminology will be automatically generated in the SPHN Dataset2RDF tool.
The hierarchy of terminologies would be intepreted in the project-specific RDF Schema as follows:
- sphn:Terminology
ATC
CHOP
…
- project:Terminology
TERMINOLOGY A
TERMINOLOGY B
…
SNOMED CT
…
Example
The Genotech project would like to create the concept of skin moisture as a measurement.
We take the sphn:Measurement as a concept that a project would like to reuse as parent for project:Skin Moisture Measurement.
general name |
general description |
type |
|
|---|---|---|---|
concept |
Measurement |
annotation used to indicate the size or magnitude of […] |
SPHNConcept |
composedOf |
result |
outcome of the concept |
Result |
composedOf |
start datetime |
start datetime of measurement |
temporal |
… |
… |
… |
… |
concept |
Skin Moisture Measurement |
hydration state of the outer epidermis |
Measurement |
inherited |
result |
outcome of the concept |
SkinMoisture |
inherited |
measurement datetime |
datetime of measurement |
temporal |
… |
… |
… |
… |
Note
in the example, we assume a SkinMoisture concept exist (or is created by the project) to encode the actual value and is a child of “Result”. Please observe that the inherited ‘result’ is reused in the Skin Moisture Measurement but we are restricting the value space to be SkinMoisture
In this example, the genotech project inherits all composedOf from Measurement under the concept Skin Moisture. The semantics are respected and the meaning of ‘inheritance’ used as it should be.
5. Defining value set or subsets
For a composedOf, a limited set of values or subset of values can be defined. This information is provided in the value set or subset column. This column must be filled according to certain rules which depends on different scenarios.
The below listed scenarios are supported in SPHN.
Scenario 1: restrict value set to a list of codes from one Terminology only, no descendants allowed
In the column standard, add the name of the Terminology (as defined in the sheet “Coding System and Version”). In the column value set or subset, provide the list of allowed codes with their label. The label is enclosed with the pipe | symbol. Each combination of code+label must be separated by a semi-colon. Code and label are separated with a space.
The pattern is:
code |label|; code |label|
Concrete example with scenario 1 standard
value set or subset
SNOMED CT
419199007 |Allergy to substance (finding)|; 782197009 |Intolerance to substance (finding)|
Scenario 2: restrict value set to a list of codes from one Terminology only, descendants allowed
In the column standard, add the name of the Terminology (as defined in the sheet “Coding System and Version”). In the column value set or subset, follow scenario 1. In addition, add in front of each code descendant of:.
The pattern is:
descendant of: code |label|; descendant of: code |label|
Concrete example with scenario 2 standard
value set or subset
SNOMED CT
descendant of: 63075001 |Monitoring of cardiac output/cardiac index (regime/therapy)|; descendant of: 117610000 |Measurement of cardiac output (procedure)|
Warning
It is currently not allowed to have value sets mixing scenario 1 and scenario 2. If such use case occurs in your project, please contact the DCC (dcc@sib.swiss). This rule also applies to the scenarios below.
Scenario 3: restrict value set to a list of codes from one Terminology but open to other terminologies or standard codes
In the column standard, add the name of the Terminology (as defined in the sheet “Coding System and Version”) followed by
or other. In the column value set or subset, start withforfollowed by the name of the Terminology, followed by a colon symbol. Then list the codes following scenario 1 or scenario 2.The pattern is:
for terminology: descendant of: code |label|(when descendants are allowed)
Concrete example with scenario 3 standard
value set or subset
SNOMED CT or other
for SNOMED CT: descendant of: 272181003 | Clinical equipment|
Scenario 4: restrict value set to a list of codes from one Terminology or codes from a standard not available in RDF (i.e. treated as a Code) only
In the column standard, add the name of the Terminology followed by
orand add the name of the standard which is not available in RDF (as defined in the sheet “Coding System and Version”). In the column value set or subset, start withforfollowed by the name of the Terminology, followed by a colon symbol. Then list the codes following scenario 1 or scenario 2.The pattern is:
for terminology: descendant of: code |label|; for standard: descendant of: code |label|(when descendants are allowed)
Concrete example with scenario 4 standard
value set or subset
SNOMED CT or EDQM
for SNOMED CT: descendant of: 736542009 |Pharmaceutical dose form (dose form)|; for EDQM: descendant of: EDQMPDF
Note
Scenario 1 is also possible in this case (i.e. no descendants allowed) but this must apply consistently to coding systems restricted. There can’t be a mix of one coding system allowing descendants and a second coding system not allowing descendants. If such case is needed for the project, please get in touch with the DCC (dcc@sib.swiss).
Scenario 5: restrict value set to a list of codes from a Terminology but possible to extend to other codes from that Terminology
In the column standard, add the name of the Terminology. In the column value set or subset, start with
extendable value setfollowed by the name of the Terminology, followed by a colon symbol. Then list the codes following scenario 1 or scenario 2.The pattern is:
extendable value set: terminology: code |label|; code |label|
Concrete example with scenario 5 standard
value set or subset
SNOMED CT
extendable value set: SNOMED CT: 29707007 |Toe structure (body structure)|; 7569003 |Finger structure (body structure)|; 48800003 |Ear lobule structure (body structure)|
Scenario 6: restrict value set to a list of set of terms provided as value set (not coming from a standard, hence not having a code)
Keep the column standard empty. In the column value set or subset, provide the list of terms in lowercase letters; separated by a semi-colon. In this case, we do not enclose the labels with the pipe symbol
The pattern is:
label; labelwhere label the human-readable term(s) defined in SPHN which will be interpreted as an individual in RDF (i.e. a possible instance value).
Concrete example with scenario 5 standard
value set or subset
principal; secondary; complementary
Scenario 7: restrict value set of a nested composedOf (property path restriction)
It may happen that you have a composedOf that has a concept as its value and you want to restrict the value space of one of the composedOf belonging to that concept (instance). In some cases the nesting can be of arbitrary length depending on how the concepts (and composedOfs) are defined.
For example, in the case of
sphn:InhaledOxygenConcentration, there is a composedOf calledhasQuantityand we want to state that theUnitused to represent theQuantityis restricted to%from UCUM. Property paths are a way to express this in an unambiguous manner in the SPHN Dataset where we are saying that the value space ofhasQuantity -> Quantity -> hasUnit -> Unit -> hasCodeis restricted to%.Following are a list of possible sub-scenarios where we use property paths and how the expression can be combined with other possible keywords in the valueset or subset column of the SPHN Dataset:
Restrict value space of a property of an instance that is the value of the current composed of
Warning
Disclaimer: Examples are for the sake of demonstration and may not reflect real use cases.
The pattern is: composedOf restricted to: code
Example with sub-scenario 7.1 concept reference
composedOf
standard
value set or subset
Access Device Presence
medical device
SNOMED CT
type code restricted to: 105789008
Restrict the value space of a nested property of an instance that is the value of the current composed of
The pattern is:
composedOf -> nested_composedOf restricted to: code-> is the symbol that denotes that the composed of succeeding the symbol is a nested composed of
Example with sub-scenario 7.2 concept reference
composedOf
standard
value set or subset
Inhaled Oxygen Concentration
quantity
UCUM
unit -> code restricted to: %
Restrict value space of a double-nested property of an instance that is the value of the current composed of
The pattern is:
composedOf -> outer_nested_composedOf -> inner_nested_composedOf restricted to: codeouter_nested_composedOf is the name of the composed of that belongs to the instance that will be in the value space of composedOf
inner_nested_composedOf is the name of the composed of that belongs to the instance that will be in the value space of outer_nested_composedOf
Hypothetical example with sub-scenario 7.3 concept reference
composedOf
standard
value set or subset
ClassXYZ
prop1
SNOMED CT
prop2 -> prop3 -> prop4 restricted to: 12345
Restrict value space of a property of an instance that is the value of the current composed of to multiple codes separated by semi-colon
The pattern is:
composedOf restricted to: code1; code2; code3
Hypothetical example with sub-scenario 7.4 concept reference
composedOf
standard
value set or subset
Circumference
body site
SNOMED CT
code restricted to: 69536005; 33673004; 29836001; 45048000
Restrict value space of a property of an instance that is the value of the current composed of to multiple codes from multiple terminologies separated by semi-colon
The pattern is:
composedOf restricted to: for terminology1: code1; code2; for terminology 2: code3; code4
Hypothetical example with sub-scenario 7.5 concept reference
composedOf
standard
value set or subset
ClassXYZ
prop1
SNOMED CT or other
prop2 restricted to: for SNOMED CT: 12345; 45353; for CHOP: Z23.1; Z43.3
Restrict value space of a property of an instance that is the value of the current composed of to descendants of a code from a terminology
The pattern is:
composedOf restricted to: descendant of: code
Example with sub-scenario 7.6 concept reference
composedOf
standard
value set or subset
Blood Pressure
body site
SNOMED CT
code restricted to: descendant of: 113257007; 40983000; 68367000
Note
Here we are saying that descendant of snomed:113257007 OR descendant of snomed:40983000 OR descendant of snomed:68367000 are allowed (i.e. the descendant of applies to ALL the codes mentioned in the value set).
6. Defining cardinalities
An important and required step during schema generation is the definition of cardinalities. Cardinalities represent a numeric relationship between two entities restricting with a minimum and a maximum the number of instances that are enabled.
For example, a schema modelling Movie, Actor and Director can state the following cardinalities:
A Movie can have 1:n (one to many) Actors
A Movie has 1:1 (one and only one) Director
Figure 3: Example of a Movie schema with cardinality definition and showcase of valid and invalid data examples.
In Figure 3, data about the Movie Avatar with two actors and one director (left) is valid according to the cardinality restrictions imposed in the schema while the data example shown with two directors (right) is invalid.
In SPHN, cardinalities are defined in the SPHN Dataset and taken into consideration in the SPHN RDF Schema. A project must therefore encode cardinalities in the SPHN Dataset Template for them to be translated into the project-specific RDF Schema when converting the project Dataset with the SPHN Schema Forge tool.
The SPHN Dataset defines for instance that a BodySite must have one and only one instance of a Code (cardinality 1:1) and it can have up to one instance of a Laterality (cardinality 0:1) as shown in Figure 4:
Figure 4: Example of the SPHN schema with cardinality definition for BodySite and showcase of valid and invalid data examples.
There are four kind of cardinalities that can be defined in SPHN:
0:1 - no instance or at most one instance of the target concept connected to an instance of the source concept
1:1 - there must be at least one and only one instance of to the target concept connected to an instance of the source concept
0:n - no instance or any number of instances of the target concept connected to an instance of the source concept
1:n - there must be at least one instance of the target concept and there can be more connected to an instance of the source concept
Cardinalities must be provided for each value of a composedOf of a concept and, when relevant, should be provided to show the connectivity between a concept and the Administrative Case, the Subject Pseudo Identifier, the Data Provider and the Source System (these four will be referred as main concepts in this section). To add these information, five columns are available at the end of the ‘Concepts’ sheet in the SPHN Dataset Template:
cardinality for composedOf: intended for the annotation of cardinalities of the value of a composedOf of a concept. Only lines where the column ‘concept or concept compositions or inherited’ is ‘composedOf’ must be filled.
cardinality for concept to Administrative Case: intended for the annotation of cardinalities related to the association of the concept to the Administrative Case
cardinality for concept to Data Provider: intended for the annotation of cardinalities related to the association of the concept to the Data Provider
cardinality for concept to Subject Pseudo Identifier: intended for the annotation of cardinalities related to the association of the concept to the Subject Pseudo Identifier
cardinality for concept to Source System: intended for the annotation of cardinalities related to the association of the concept to the Source System
Thinking to apply when defining cardinalities
When defining your cardinalities, you should think what can be logically linked.
The definition of cardinalities can follow some rules:
composedOf cardinalities need to be made mandatory (must be 1:1 or 1:n ) when that information is crucial for the concept. The thinking should be: would this concept make sense without this value? (e.g. a measurement without a value do not make sense, a patient without an identifier does not make sense)
and logically, composedOf cardinalities which are not mandatory must be 0:1 or 0:n
composedOf which are pointing to a datetime must be either 0:1 or 1:1: a concept has at most once a given datetime (e.g. an instance of Body Height has only one measurement datetime. A patient can have their body height measured multiple times - so there are multiple date times and values - but each measurement is a different instance of Body Height where each instance has one value and one measurement datetime.)
concepts with a datetime composedOf (e.g. measurements, procedures, diagnosis) are specific to a patient. Therefore, a single instance of such concepts can be connected at most to one single patient (i.e. Subject Pseudo Identifier). The cardinality of such a concept to a Subject Pseudo Identifier would be 1:1
If your concepts inherits from an SPHN concept, the cardinalities of the inherited composedOfs and the cardinalities to the three main concepts must either be the same as defined in the SPHN Dataset or it can only be narrower. If you need to widen it, then a change request might be needed in the SPHN Dataset before you could apply such wider cardinalities for your concept, please contact the DCC for such case
some concepts can have instances reusable in multiple context (e.g. Birth Date, Death Date). These concepts can have a cardinality of 0:n or 1:n to the three main concepts. For example, there can be multiple patients (Subject Pseudo Identifier) born on the 24 January 1997 at 21:30. That instance of Birth Date, provided in the data with the same identifier (IRI), can be used in multiple patients
Sometimes, we need to be mindful of specific use cases or situations. A few of such situations are:
concepts often come with an Administrative Case in the setting where the data is provided by a hospital. However, it is good to keep in mind that in another setting, there might not be an Administrative Case (e.g. cohort, registries). Therefore, when assigning the cardinality of a concept to an Administrative Case, consider this detail and don’t necessarily always make it 1:1 but rather 0:1
some concepts can have no connection to any of the three main concepts, in which case the assumption is that the concept must be reused as a value of a composedOf of another concept
In case of doubt or special cases, do not hesitate to contact the DCC (dcc@sib.swiss).
7. Transform the Dataset Template to a RDF Schema
Once the Dataset Template is filled, a last step remains to generate the project-specific RDF Schema and (if wanted) all related content from SPHN Semantic Interoperability Framework (SHACL rules, SPARQL queries, pyLODE Schema Visualization). The updated Dataset Template, which now has become the project-specific Dataset can be given as input to the SPHN Schema Forge (https://schemaforge.dcc.sib.swiss), a web service that will generate all the previously cited materials.
Note
The project can add sheets to the Dataset Template for keeping track of additional metadata (e.g., release notes) but these information will not be processed by the SPHN Schema Forge.
7.1 Common mistakes translated as errors in the SPHN Schema Forge
The following table provides an overview of the possible error messages provided by the Dataset2RDF tool (integrated in the SPHN Schema Forge) when translating the Dataset Template to an RDF Schema and how to interpret them.
Error log
How to interpret the error
[Row XXX] Project concept ‘XXX’ occurs more than once (Row XXX, Row XXX)
You can’t have two lines with the same project concept name. You must delete one of the lines (and check the composedOfs that may be associated).
[Row XXX] Project concept ‘XXX’ should have ‘XXX’ as its parent instead of “SPHNConcept”
You can’t have a project concept name whose parent is SPHNConcept. You have to use the root concept name of your project (project:<PROJECT>Concept).
[Row XXX] Project composedOf ‘XXX’ should have ‘XXX’ as its parent instead of ‘SPHNAttributeObject’
A project composedOf (which therefore does not exist in SPHN) can’t have as parent the SPHNAttributeObject. Note that this does not apply to SPHN composedOf that may be reused in a project concept as a composedOf.
[Row XXX] Inherited composedOf ‘XXX’ has cardinality which is broader (XXX) than what is expected (XXX)
The inherited composedOf has a broader cardinality when comparing to the same composedOf defined in the parent concept. The cardinality must either be the same as in the parent concept or be narrower.
[Row XXX] composedOf ‘XXX’ has cardinality with is unrecognized (XXX) when compared to what is expected (XXX)
The composedOf is most likely coming from an inherited concept and its cardinality has been changed but the cardinality pattern is incorrect.
[Row XXX] Project composedOf ‘XXX’ should have ‘XXX’ as its parent instead of ‘XXX’
The parent given to a project composedOf appears to be wrong (typo, wrong concept).
[Row XXX] inherited project composedOf ‘XXX’ should have ‘XXX’ as its parent instead of ‘XXX’
The parent provided to a project inherited composedOf appears to be wrong (typo, wrong concept).
[Row XXX] inherited project composedOf ‘XXX’ has a parent ‘XXX’ that does not exist
The project inherits a composedOf but the parent in the ‘parent’ column does not exist within the project.
[Row XXX] composedOf ‘XXX ‘ has type (XXX) which is not in the same scope when comparing with composedOf ‘XXX’ with type (XXX) in parent concept ‘XXX’
A concept which is a child of another concept is defined. One of the inherited composedOf of that child concept differs in the type compared to the same composedOf defined in the parent. Make sure to align the types of these composedOfs, which must be the same.
[Row XXX] inherited SPHN composedOf ‘XXX’ should have ‘XXX’ as its parent instead of ‘XXX’
The composedOf which is inherited in the project concept comes from SPHN. Therefore, the parent must be the same as defined in the SPHN composedOf.
[Row XXX] Concept ‘XXX’ has ‘cardinality for concept to XXX’ which is broader (XXX) than its parent (XXX)
The project concept in question is inheriting from another parent (SPHN or project-specific) concept. However the cardinality applied to one of the core concepts (Subject Pseudo Identifier, Source System, Administrative Case or Data Provider) is broader than the one defined in the parent concept. This is not allowed. The cardinality must either be the same or can only be narrower.
[Row XXX] Concept ‘XXX’ has ‘cardinality for concept to XXX’ which is unrecognized (XXX) when compared to its parent (XXX)
The project concept in question is inheriting from another parent (SPHN or project-specific) concept. However the cardinality applied to one of the core concepts (Subject Pseudo Identifier, Source System, Administrative Case or Data Provider) is not recognizable when compared to the parent. There is some formatting issue with the cardinality expression.
[Row XXX] SPHN concept ‘XXX’ has an invalid IRI ‘XXX’. SPHN concept should have SPHN IRI.
The project extends an SPHN concept but adds a project-IRI for this SPHN concept. This is not allowed. You must use SPHN IRI for SPHN concepts even if they are extended for a project.
[Row XXX] Project concept ‘XXX’ has an invalid IRI ‘XXX’. Project concept should have project IRI
The project defines a project concept but uses the SPHN IRI for the concept. This is not allowed. You must use a project-specific IRI.
[Row XXX] SPHN composedOf ‘XXX’ has an invalid IRI ‘XXX’. SPHN composedOf should have SPHN IRI
The project is reusing an existing SPHN composedOf (i.e. the general name does not contain a project prefix) but modified the IRI (i.e. most likely with the project-specific IRI). This is not allowed. You must use the same IRI as defined in SPHN.
[Row XXX] Project composedOf ‘XXX’ has an invalid IRI ‘XXX’. Project composedOf should have project IRI
The project defines a composedOf but uses the SPHN IRI for the composedOf. This is not allowed. You must use a project-specific IRI.
[Row XXX] composedOf ‘XXX’ has a standard ‘XXX’ that is not allowed since parent composedOf ‘XXX’ allows only OBI
The standard column of the inherited composedOf specifies the name of a terminology which is not specified or allowed in the parent’s composedOf.
[Row XXX] inherited SPHN composedOf ‘XXX’ has a reference ‘XXX’ that does not exist
The reference column of the inherited composedOf appears to be wrong (typo, wrong concept).
[Row XXX] inherited project composedOf ‘XXX’ has a reference ‘XXX’. The reference concept inherits from concept ‘XXX’ that does not exist
The inherited composedOf has a reference concept. This reference concept has a parent defined in the ‘parent’ column that does not exist.
[Row XXX] inherited project composedOf ‘XXX’ has a reference ‘XXX’. The reference concept does not have a ‘type’ defined
The inherited project composedOf has a reference concept. This reference concept does not have a ‘type’ defined in the ‘type’ column.
[Row XXX] inherited project composedOf ‘XXX’ has a reference ‘XXX’ that does not exist
The inherited project composedOf has a reference concept that does not exist.
[Row XXX] composedOf ‘XXX’ has type (XXX) which is not in the same scope when comparing with composedOf ‘XXX’ with type (XXX) in parent concept ‘XXX’
The type column of the composedOf appears to be wrong (typo, wrong concept).
[Row XXX] inherited SPHN composedOf ‘XXX’ has a parent ‘XXX’ that does not exist
The parent column of the inherited composedOf appears to be wrong (typo, wrong concept).
[Row XXX] inherited SPHN composedOf ‘XXX’ does not exist in parent concept ‘XXX’
A concept defines a composedOf as being inherited but this composedOf doesn’t seem to exist in the parent concept. This can be due to a typo, wrong composedOf name or this can be fixed by changing the column ‘concept or concept compositions or inherited’ from ‘inherited’ to ‘composedOf’.
[Row XXX] inherited project composedOf ‘XXX’ does not exist in parent concept ‘XXX’
A project concept defines a project composedOf as being inherited but this composedOf doesn’t seem to exist in the parent concept. This can be due to a typo, wrong composedOf name or this can be fixed by changing the column ‘concept or concept compositions or inherited’ from ‘inherited’ to ‘composedOf’
[Row XXX] inherited SPHN composedOf ‘XXX’ should have ‘SPHNAttributeObject’ as its parent instead of ‘XXX’
The project uses an inherited SPHN composedOf but changes its parent from SPHNAttributeObject to a project-specific attribute object. This is not allowed.
[Row XXX] Concept ‘XXX’ is repeated but with different values for column ‘XXX’
The SPHN concept, which is extended by the project, has been defined with a different value in a given column. This is not allowed, the column in question should be the same as the one defined initially in the SPHN concept.
[Row XXX] Repeated concept ‘XXX’ has a composedOf ‘XXX’ with different values for column ‘XXX’
The SPHN concept, which is extended by the project, has been defined with an already defined SPHN composedOf but some values have changed. This is not allowed. In repeated SPHN concepts, the project must create its own prefixed composedOfs (even if the general name of the composedOf is the same as the one defined in the SPHN concept).
[Row XXX] Project composedOf ‘XXX’ has type ‘qualitative’. Either ‘valueset or subset’ column should be defined OR the parent should be an ‘Attribute Datatype’
The project composedOf has an attribute object as its parent and the type as ‘qualitative’. If this is truly the case then there should be a valueset defined in the ‘valueset or subset column’. If there are no valuesets then this property’s parent should be an Attribute Datatype.
[Row XXX] SPHN concept ‘XXX’ should have ‘XXX’ as its parent instead of ‘XXX’
The project extends an SPHN concept but uses a project root concept as its parent. This is not allowed. SPHN concept must have the SPHN root concept as its parent.
[Row XXX] Project concept ‘XXX’ should have ‘XXX’ as its parent instead of ‘XXX’
The project defines a concept whose parent is the SPHN root concept instead of project root concept. This is not allowed. Project concepts must have the project root concept as its parent OR a project concept OR an SPHN concept (excluding the SPHN root concept).
[Row XXX] Cannot find parent ‘XXX’ for concept ‘XXX’
Cannot find the parent defined in the ‘parent’ column for a defined concept.
[Row XXX] SPHN composedOf ‘XXX’ has a parent ‘XXX’ that does not exist
The project reuses an SPHN composedOf and redefines the parent of this composedOf but the parent does not exist.
[Row XXX] Project concept ‘XXX’ has parent ‘XXX’ with type ‘XXX’; If parent is a root concept then type should remain empty
The project defines a concept where the parent in the ‘parent’ column refers to a root concept but the type in the ‘type’ column refers to another concept. This is not allowed. When the ‘parent’ column refers to a root concept, the ‘type’ column should remain empty.
[Row XXX] Project concept ‘XXX’ has parent ‘XXX’ with type ‘XXX’; Type and parent should always refer to the same concept
The project defines a concept where the parent in the ‘parent’ column refers to a concept but the type in the ‘type’ column refers to another concept. This is not allowed. The ‘parent’ and ‘type’ column should always refer to the same concept when the ‘parent’ is not a root concept.
[Row XXX] SPHN composedOf ‘XXX’ does not exist. If this is a project composedOf then the name should have a project prefix
The project reuses a composedOf that does not exist in SPHN. If the composedOf is a project composedOf then the name in the ‘general concept name’ column should include the project prefix.
[Row XXX] SPHN composedOf ‘XXX’ should have SPHNAttributeObject as its parent instead of ‘XXX’. If this is a project composedOf then the name should have a project prefix
The project reuses an SPHN composedOf but redefines its parent as project attribute object. This is not allowed. SPHN composedOf should have SPHNAttributeObject as its parent. If the composedOf is to be a project composedOf then the name in the ‘general concept name’ column should include the project prefix.
[Row XXX] Project composedOf ‘XXX’ has type ‘XXX’. Thus its parent should be an Attribute Datatype
The project defines a composedOf with a specific type. But the parent in the ‘parent’ column is inconsistent with the defined type and should actually be a project-specific attribute datatype.
[Row XXX] Project composedOf ‘XXX’ has type ‘XXX’. Thus its parent should be an Attribute Object
The project defines a composedOf with a specific type. But the parent in the ‘parent’ column is inconsistent with the defined type and should actually be a project-specific attribute object.
Found missing column name(s): XXX
One (or more) columns are missing from the Dataset Template.
Note
This is not an exhaustive list of errors that may be displayed in the SPHN Schema Forge. In case of misunderstanding with an obtained error message, please contact dcc@sib.swiss.
Option 2: Generate an RDF Schema using an ontology editor
The project also has the possibility to generate their RDF Schema, compliant with SPHN using an editor of their choice (e.g. Protégé). This option can be considered for experienced users with Semantic Web Technologies.
Note
A tutorial based on previous versions of the SPHN RDF Schema can be found here: Expanding the SPHN RDF Schema Please note that some of the statements of the videos might not be valid anymore.
In this case, the project could still run the SPHN Schema Forge (https://schemaforge.dcc.sib.swiss) to obtain the different files (SHACLs, HTML, SPARQLs) related to their project schema.
Reporting back to DCC
The DCC welcomes any feedback to the SPHN Dataset and to the SPHN RDF Schema to improve these specifications. For any change requests to the SPHN Dataset (or SPHN RDF Schema), please use the adequate concept template for reporting the request and submit the document to dcc@sib.swiss.