External terminologies
Note
To find out more watch the Semantic standards training
SPHN provides the external terminologies via the Terminology Service in RDF format. Currently, the following terminologies are provided:
ATC (including historical version since 2016)
CHOP (including historical version since 2013)
ICD-10-GM (including historical version since 2013)
LOINC (including historical version since version 2.69)
SNOMED CT (including historical version since 2021-01-31; note that only the December release of the Swiss Edition is provided)
If you are missing a terminology important for your project, write a request to dcc@sib.swiss.
ATC
The Anatomical Therapeutic Classification (ATC, https://www.whocc.no/atc_ddd_index/) is a drug classification that categorizes active substances of drugs according to the organ or system where their therapeutic effect occurs. ATC is provided by the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC) and is updated once a year. In Switzerland, the ATC code is used by Swissmedic (https://www.swissmedic.ch) in the extended list of medicinal products approved by Swissmedic.
The hierarchy of ATC is divided in five levels of classification (see Figure 1):
Level 1: Anatomical main group (represented by one letter)
Level 2: Therapeutic subgroup (represented by two digits)
Level 3: Pharmacological subgroup (represented by one letter)
Level 4: Chemical subgroup (represented by one letter)
Level 5: Chemical substance (represented by two digits)
Figure 1. Example of the ATC code A02AB03. The different levels and their meaning (label) are presented for the ATC code A02AB03 which corresponds to the aluminium phosphate drug.
ATC is used as one recommended standard for the SPHN concept: Substance (https://www.biomedit.ch/rdf/sphn-schema/sphn#Substance).
Information for use in data science
The following information is taken from the 2021 Guidelines that are available on the ATC Website https://www.whocc.no/atc_ddd_index/.
Many medicines are used and approved for two or more indications, while normally only one ATC code will be assigned. Besides, ATC codes are often assigned according to the mechanism of action rather than therapy. An ATC group may therefore include medicines with many different indications, and drugs with similar therapeutic use may be classified in different groups.
A medicinal substance can be given more than one ATC code if it is available in two or more strengths or routes of administration with clearly different therapeutic uses (e.g. Finasteride).
Implementation in RDF for SPHN
The ATC code (English version) made available by WHOCC in an Excel file has been translated into an RDF representation via a Python script.
Since ATC is not providing a generic code for grouping all ATC codes together,
an ATC class has been created using the namespace <https://biomedit.ch/rdf/sphn-resource/atc/> (see Figure 2).
A version IRI is provided for each version of ATC in RDF (e.g., https://biomedit.ch/rdf/sphn-resource/atc/2021/1/),
which is composed of the namespace followed by the year of release of ATC and the version
of the RDF generated for that release.
The ATC class puts together ATC codes in the RDF provided by WHOCC (see again Figure 2).
These ATC codes use the namespace following the ATC website link: <https://www.whocc.no/atc_ddd_index/?code=>,
enabling the codes to be dereferencable on the web. When data is referring to an ATC code, it must use this link as namespace.

Figure 2. Grouping of ATC codes under the ‘ATC’ class. All ATC codes are grouped under the SPHN defined class ATC (https://biomedit.ch/rdf/sphn-resource/atc/ATC).
In the ATC RDF, three types of information are stored (see Figure 3):
The ATC code itself, which is defined as a class,
The name (label) of the ATC code, represented with the use of the property
rdfs:label,The hierarchy of the codes, represented with the use of the property
rdfs:subClassOf(see Figure 4). Level 1 defines the main classes. Level 2 codes are sub-classes of Level 1. Level 3 are sub-classes of Level 2. Level 4 are sub-classes of Level 3. Level 5 are sub-classes of Level 4.
Figure 3. RDF representation of an ATC code. A code is defined as an RDF Class and is associated with a specific label. In addition, a code A can be linked to another code B as a subclass of the latter (i.e., code A is a child of code B and code B is a parent of code A).
Figure 4. Representation of the relationship between ATC codes in RDF. The property subClassOf defines whether a specific ATC code is a child of another ATC code. For instance, level 5 ATC codes are defined as sub-classes of level 4 ATC codes and level 4 ATC codes are sub-classes of level 3 ATC codes. With the reasoning possibilities offered in RDF, one can infer that a level 5 ATC code is a sub-class of a level 1 ATC code.
The following information provided by ATC are not incorporated in RDF, either because they are not relevant or are reflected in other concepts of SPHN:
The DDD (average maintenance dose per day of a substance)
The Unit (unit of the dose)
The Administration route (route of administration of the substance)
The Comment (additional comment about a substance).
Finally, the versioning strategy is applied to ATC RDF files meaning that each RDF file contains codes from the latest version, versioned IRIs of previous versions of ATC back until version 2016. More information on the versioning
Usage rights
The copyright follows the instructions provided by WHO regarding ATC (https://www.whocc.no/copyright_disclaimer/): “Use of all or parts of the material requires reference to the WHO Collaborating Centre for Drug Statistics Methodology. Copying and distribution for commercial purposes is not allowed. Changing or manipulating the material is not allowed.”
CHOP
The “Schweizerische Operationsklassifikation” (CHOP) is a Swiss medical coding system for procedures performed in hospitals. CHOP covers a wide range of procedures including surgical, radiotherapeutic, diagnostic procedure such as ECGs, and procedures for patient activation (e.g. walking up and down stairs) as well as planning activities. CHOP, initially based on the ICD-9-CM, is a Swiss classification used mainly for billing purposes. CHOP versions are released annualy by the Federal Statistical Office (FSO) and are free to download. The CHOP classification is available in German, French and Italian.
The hierarchy of CHOP is divided into six levels of classification (see example in Figure 5):
The code of the first level starts with a “C” followed by a number and corresponds to the chapter.
The code of each of the other levels starts with either a number or a letter.
Figure 5. Example of CHOP hierarchy. The code C0 represents the most generic level of information, corresponding to a chapter while the code 00.12.00 is the most specific level of information. The different levels enable building up the hierarchy of the classification: the lower the level; the more specific the information.
Information for use in data science
CHOP classifies procedures into one of 19 different categories (chapters).
The chapters are largely differentiated according to medical specialty.
For example, chapter C1 contains procedures on the nervous system, chapter C2 contains procedures on the endocrine system.
Chapter C16 contains various diagnostic and therapeutic procedures across all therapeutic areas,
e.g. radiography of the spine (87.2) and CT of the thorax (87.41).
Chapter C17 contains measurement instruments, e.g. scores such as mobility scores.
Chapter 18 is dedicated to rehabilitation, and chapter C0 contains procedures and interventions not classifiable elsewhere.
The hierarchies can support data aggregation but, depending on the scope of aggregation,
several chapters might need to be taken into account.
Implementation in RDF for SPHN
The CHOP coding system is available as a multi-language Excel file in which the correspondence between the German, French and Italian versions are presented. This file has been used (saved in Excel format) to generate the RDF representation via a Python script.
The namespace used in the RDF is: <https://biomedit.ch/rdf/sphn-resource/chop/>.
A version IRI is provided for each version of CHOP in RDF (e.g., https://biomedit.ch/rdf/sphn-resource/chop/2020/1/), composed of the namespace followed by the year of release of CHOP and the version of the RDF generated for that release.
In the RDF version generated of CHOP, three types of information are stored:
The CHOP code itself, which is defined as a class (
rdfs:class).The name (label) of the CHOP code, represented by the property
rdfs:labelcorresponding to the row where the item type is “T” (here “T” stands for “title”). The label is described in each of the three languages with the suffix@DEfor German,@FRfor French and@ITfor Italian.The hierarchy of the codes, represented by the property
rdfs:subClassOf.
The following information provided by CHOP are not currently incorporated in SPHN CHOP RDF:
Item type B, I, N, S and X
Codable code
Indent level
Laterality
CHOP versions from 2016 to 2022 have been translated into RDF.
Usage rights
The copyright follows the instructions provided by FSO, Neuchâtel 2022: “Wiedergabe unter Angabe der Quelle für nichtkommerzielle Nutzung gestattet” (i.e., Reproduction is authorized, except for commercial purposes, if the source is acknowledged).
ECO
The Evidence & Conclusion Ontology (ECO) is an ontology used to represent scientific evidence in biological research. Documenting the evidence that supports a scientific assertion is essential and fundamental to the scientific method. ECO provides a vocabulary that can be used to achieve this. ECO also provides a practical means by which to employ quality control measures when importing data into databases. One can also draw inferences about one’s confidence in conclusion by looking at associated evidence.
ECO describes evidence arising from laboratory experiments, computational methods, curator inferences, and other means. Many of the largest biological and genomic databases use ECO for summarizing evidence in scientific investigations.
Information for use in data science
ECO is an ontology used for representing provenance and evidence information about data points. This enhances the quality of the data so that when analyzing it, the data user can make informed decisions based on the provenance and the quality of the evidence that is given.
For instance, ECO can be used to represent information about whether a data point was computed, manually curated, obtained from a document, experimentally derived or inferred.
Implementation in RDF for SPHN
ECO is made available as-is by the SPHN DCC.
The namespace used is: http://purl.obolibrary.org/obo/ECO_
A versionIRI is provided for each version of ECO in RDF which indicates the version (or release) of ECO.
For example, http://purl.obolibrary.org/obo/eco/releases/2023-09-03/eco.owl indicates that the
ontology is from a 2023-09-03 release of ECO.
In ECO, a concept is defined with the following structure:
ECO:0000203 a owl:Class ;
rdfs:label "automatic assertion"^^xsd:string ;
IAO:0000115 "An assertion method that does not involve human review."^^xsd:string ;
oboInOwl:created_by "mchibucos"^^xsd:string ;
oboInOwl:creation_date "2010-03-18T12:36:04Z"^^xsd:string ;
oboInOwl:hasAlternativeId "ECO:00000067"^^xsd:string ;
oboInOwl:hasOBONamespace "eco"^^xsd:string ;
oboInOwl:id "ECO:0000203"^^xsd:string ;
rdfs:comment "An automatic assertion is based on computationally generated information that is not reviewed by a person prior to making the assertion. For example, one common type of automatic assertion involves creating an association of evidence with an entity based on commonness of attributes of that entity and another entity for which an assertion already exists. The commonness is determined algorithmically."^^xsd:string ;
rdfs:subClassOf ECO:0000217 ;
owl:disjointWith ECO:0000218 .
Usage rights
ECO is accessible in the public domain under CC0 1.0 Universal (CC0 1.0) License (https://www.evidenceontology.org/about_eco/#license).
EDAM
The Ontology of bioscientific data analysis and data management (EDAM, https://edamontology.org/page) is an ontology that includes terms related to data analysis and data management in life sciences such as:
Topicthat refer to the field of study of the data (e.g. biology, physics, chemistry);
Formatthat refer to computer file formats that structure the data (e.g. CSV, JSON, RDF/XML);
Operationthat refer to kinds of analysis performed on the data (e.g. Gene regulatory network analysis, pathway visualization)
Information for use in data science
EDAM is used for annotation of information related to tools, workflows, data formats in which data is encoded. It can also enable the annotation of provenance metadata in some cases.
In the case of SPHN, EDAM is currently used to refer to:
formats of data files that may be provided with the RDF graph and which contain results of specific concepts
analysis done in specific processes and data analysis, mainly related to omics data.
Implementation in RDF for SPHN
EDAM is made available as-in by the SPHN DCC.
The namespace used is: http://purl.obolibrary.org/obo/edam#
Note
For the SPHN RDF Schema 2024.2 release, the version of EDAM used is the 1.25. The SPHN DCC added a versionedIRI to the EDAM RDF file since it was missing but needed for our imports into the SPHN RDF Schema. In future releases of EDAM, the versionedIRI should be directly encoded in EDAM by the providers of that terminology.
Usage rights
EDAM is a community-built ontology. Contribution are welcome on GitHub (https://github.com/edamontology/edamontology). EDAM is available under Creative Commons Attribution 4.0 International (CC BY 4.0) license.
EFO
The Experimental Factor Ontology (EFO) provides a systematic description of many experimental variables available in EBI databases, and for projects such as the GWAS catalog. It combines parts of several biological ontologies, such as UBERON anatomy, ChEBI chemical compounds, and Cell Ontology. The scope of EFO is to support the annotation, analysis and visualization of data handled by many groups at the EBI. But EFO is also open to adding terms from external users when requested.
Information for use in data science
EFO is primarily used to annotate biomedical data from the EBI’s databases like the BioSamples database, Atlas of Gene Expression, ArrayExpress, and the GWAS Catalog. Given that EFO is an application ontology, there are several terms represented in EFO that can be used for a wide variety of use cases. In the case of SPHN, EFO is used to refer to metadata pertaining to genomics. For example, describing the nature of Assay and Sequencing Assay.
Implementation in RDF for SPHN
EFO is made available as-in by the SPHN DCC.
The namespace used is: http://www.ebi.ac.uk/efo/EFO_
A versionIRI is provided for each version of EFO in RDF which indicates the version (or release) of EFO.
For example, http://www.ebi.ac.uk/efo/releases/v3.61.0/efo.owl indicates that the ontology is from
the 3.61.0 release of EFO.
In EFO, a concept is defined with the following structure:
efo1:EFO_0005684 a owl:Class ;
rdfs:label "RNA-seq of coding RNA from single cells" ;
IAO:0000115 "An assay in which sequencing technology (e.g. Illumina) is used to generate RNA sequence, from the presumed coding transcibed regions of the genome, or analyse these or to quantitate transcript abundance in individual cells instead of a population of cells." ;
IAO:0000117 "Dani Welter" ;
rdfs:subClassOf [ a owl:Restriction ;
owl:onProperty OBI:0000293 ;
owl:someValuesFrom efo1:EFO_0007831 ],
efo1:EFO_0003738,
efo1:EFO_0008913 .
Usage rights
EFO is an application ontology primarily developed for knowledege organization and representation at EMBL-EBI. EFO is available under Apache License, Version 2.0 with terms of use.
EMDN
Introduction to the classification
The European Medical Device Nomenclature (EMDN) is a medical device classification that provides information on the types of medical devices, opposed to product identifier systems with very fine-grained distinction on the level of manufacturer and version or model of a medical device. EMDN is based on the Classificazione Nazionale Dispositivi medici (CND), the Italian nomenclature for medical devices and has been established to support the European database on medical devices (EUDAMED). At the beginning of May 2021, the first version of EMDN was released and manufacturers will be asked to use the EMDN for the registration of medical devices in EUDAMED. An EMDN code will be assigned to each Unique Device Identifier – Device Identifier (UDI-DI) in EUDAMED and the combination of UDI-DI and the associated EMDN code therefore link a product and a type identifier.
The hierarchy of EMDN is divided into seven levels of classification (see example in Figure 6):
Level 1 indicates the category of medical device, referred with a letter
Level 2 indicates the group of medical devices, referred with two numbers
Level 3 to Level 7 indicate the type of medical device, referred with a series of numbers
Figure 6. Visual representation of EMDN codes and their hierarchy.
Category W (in vitro diagnostic medical devices) is of particular interest for research in laboratory diagnostics, as it comprises types of laboratory analyzers, test kits and accessories for in vitro diagnostic devices and tools.
EMDN is used as one of the recommended standard for the SPHN concept:
Lab Analyzer (https://www.biomedit.ch/rdf/sphn-schema/sphn#LabAnalyzer).
Information for use in data science
The progressive spread and increasing use of LOINC-codes, for example, for clinical observations and laboratory tests, is an enormous benefit for multi-center projects as it increases comparability and interoperability of clinical data. Despite their advantages LOINC often provide no or rather high-level classification on the method employed to generate a laboratory result. Such information, including the type of the medical device(s) used, however, is of major interest for clinical staff and researchers.
Example 1:
EMDN code W02020201 is assigned to coagulometers, and the codes W0202020101 and W0202020102 one level below allow to distinguish semi-automated and automated methods.
Example 2:
EMDN code W0101060101 is assigned to glucose test strips. The hierarchical nature of the EMDN additionally reveals that it is blood test strips (as opposed to, e.g., urine test strips) for rapid and point of care testing.
A type categorization system like EMDN (but also others, e.g., GMDN) can be helpful where different, but similar models of medical devices (e.g. of a model series) are used for analysis in different hospitals, i.e. in cases where a distinction on the level of model adds no further benefit.
Implementation in RDF for SPHN
COMING SOON
Usage rights
EMDN is provided by the European Union.
EMDN is licensed under the Creative Commons Attribution 4.0 International (CC BY 4.0) licence.
GENO
The Genotype ontology (GENO, https://github.com/monarch-initiative/GENO-ontology/) provides a graph-based model for the representation of genetic variations described in genotypes implemented in OWL2. The ontology allows a thorough description of genotype components, relationships and characteristic (e.g., genetic variant features) found in human and other model organism, enabling the aggregation and analysis of genotype-to-phenotype (G2P) data from different sources.
The GENO ontology is developed within the scope of the Monarch Initiative (https://monarchinitiative.org), whose goal is to develop tools that benefits precision medicine, disease modeling and the exploration of genotype-environment-phenotype interactions. The ontologies developed under the umbrella of the Monarch Initiative, such as the Human Phenotype Ontology (HPO), maintain a high degree of interoperability with GENO.
GENO also reuses several terms (and relationships) from Sequence Ontology and extends it further to describe about variants and thus enabling better descriptions of genetic variation data.
Information for use in data science
At its core, the GENO ontology relies on a representation of the genotype reduced to its essential components. By doing so, a complex genotype can be summarized by a combination of its genetic variations starting from the full genotype down the specific alleles and sequences alterations as seen in Figure 7.
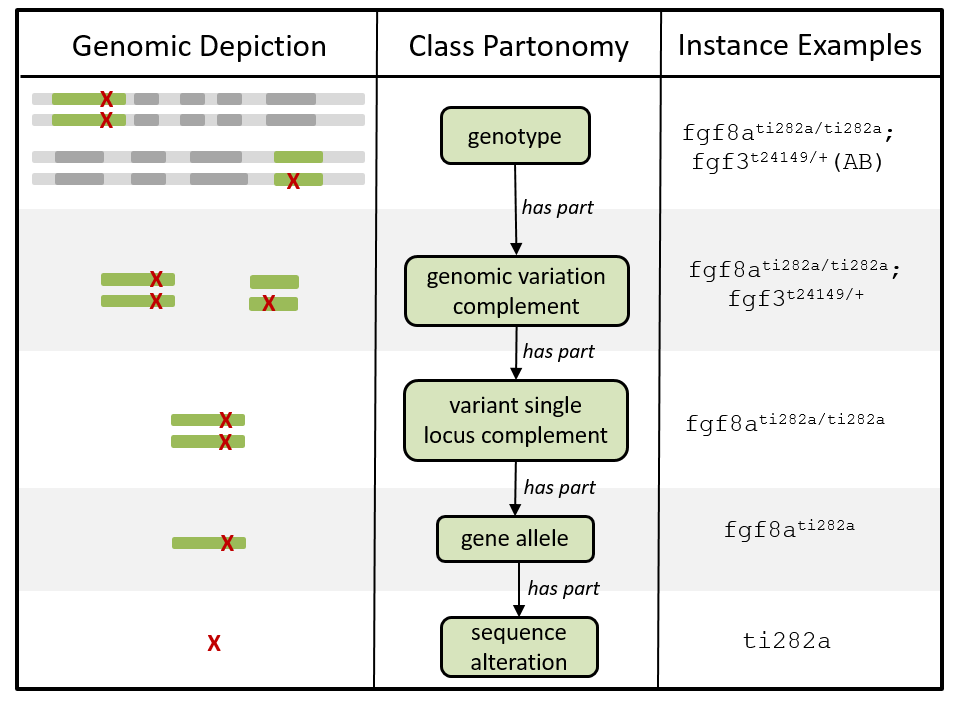
Figure 7. Genotype decomposition in GENO. The genotype can be represented at the highest level (total genomic changes across the entire genome) down to its subcomponents such as multi-loci variations, single-locus variations and single alleles (source: https://github.com/monarch-initiative/GENO-ontology).
The concept of genotype in GENO is divided into three categories:
Intrinsic genotypes or variation within the sequence.
Extrinsic genotypes or variation in gene expression.
Effective genotypes: sum of intrinsic and extrinsic genotypes. Equal to the total variation in gene sequence or expression.
This broad definition of genotypes featured by GENO allows to annotate the classing sequence-level variations that lead to a certain phenotype as well as expression level variations caused by external agents such as RNA interference constructs.
In addition to genotypes, GENO borrows from other Monarch’s ontologies to allows the description of experimental tools and reagents used for the generation genotype data. By using the GENO ontology, research projects are able to annotate genotype information in a structured way, enabling the integration of G2P data across diverse systems and laying the logical foundation for analysis and inference between phenotypes and variants.
Implementation in RDF for SPHN
GENO is made available as-is by the SPHN DCC.
The namespace used is: <http://purl.obolibrary.org/obo/GENO_>
A version IRI is provided for each version of GENO in RDF which indicates the version (or release) of GENO.
For example, http://purl.obolibrary.org/obo/geno/releases/2022-08-10/geno.owl indicates
that the ontology is from a 2022-08-10 release of GENO.
In GENO, a concept is defined with the following structure:
GENO:0000054 a owl:Class ;
rdfs:label "homo sapiens gene" ;
IAO:0000115 "A gene that originates from the genome of a homo sapiens." ;
rdfs:subClassOf [ a owl:Restriction ;
owl:onProperty RO:0002162 ;
owl:someValuesFrom NCBITaxon:9606 ],
SO:0000704 ;
owl:equivalentClass [ a owl:Class ;
owl:intersectionOf ( SO:0000704 [ a owl:Restriction ;
owl:onProperty RO:0002162 ;
owl:someValuesFrom NCBITaxon:9606 ] ) ] .
Usage rights
GENO is published by the Monarch initiative and is an open-source ontology, implemented in OWL2 under a Creative Commons 4.0 BY license. (https://github.com/monarch-initiative/GENO-ontology/)
GenEpiO
Genomic Epidemiology Ontology (GenEpiO) covers vocabulary necessary to identify, document and research foodborne pathogens and associated outbreaks. It covers various subdomains including genomic laboratory testing, specimen and isolate metadata, and epidemiological case investigations. It is an application ontology that draws on many other ontologies including anatomy, taxonomy, disease, symptoms, environment and food types for foodborn pathogen metadata.
Information for use in data science
GenEpiO is an application ontology with the goal of providing a single, open-source, and accessible set of terms to use in databases and user interfaces. GenEpiO makes use of OWL to define and organize terms. It also relies on the hierarchy from Basic Formal Ontology (BFO) and Ontology for Biomedical Investigations (OBI) to structure and organize terms in GenEpiO.
In the context of SPHN, GenEpiO is used to represent metadata pertaining to genomics. For example, describing the type of sample library preparation for Sequencing Assays.
Implementation in RDF for SPHN
GenEpiO is made available as-is by the SPHN DCC.
The namespace used is: http://purl.obolibrary.org/obo/GENEPIO_
A versionIRI` is provided for each version of GenEpiO in RDF which indicates the version (or release) of GenEpiO.
For example, http://purl.obolibrary.org/obo/genepio/releases/2023-08-19/genepio.owl indicates that the
ontology is from a 2023-08-19 release of GenEpiO.
In GenEpiO, a concept is defined with the following structure:
GENEPIO:0000061 a owl:Class ;
rdfs:label "AccuProbe Listeria monocytogenes culture identification reagent kit"@en ;
IAO:0000114 IAO:0000122 ;
IAO:0000115 "An AccuProbe reagent kit for identifying Listeria monocytogenes."@en ;
IAO:0000117 "Damion Dooley"@en ;
IAO:0000119 "URI: http://www.hologic.com/sites/default/files/package%20inserts/103051F-EN-RevC.pdf" ;
oboInOwl:inSubset "GENEPIO" ;
rdfs:subClassOf OBI:0001879 .
Usage rights
GenEpiO is accessible in the public domain under the CC BY 3.0 license (https://github.com/GenEpiO/genepio/blob/master/LICENSE).
HGNC
The Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) is the global authority that assigns standardized nomenclature to human genes. All approved symbols are complemented with additional information such as gene groups, genomic, proteomic and phenotypic information and are stored and accessible within a curated online repository (http://genenames.org). The HGNC database is updated on monthly basis.
The nomenclature provides a symbol and a name for protein-coding genes, pseudogenes that retain significant homology to a functional ancestor, non-coding RNA genes and functional read-through transcripts that have been previously annotated. Importantly, the following cases are not covered by the HGNC nomenclature:
Sequence-variant nomenclature: This issue is the responsibility of the Human Genome Variation Society (HGVS)
Product of gene translocations or fusions: No official naming guidelines exist at the moment. HGNC issued a recommendation for the use of the SYMBOL1/SYMBOL2 format.
Protein nomenclature: The naming of proteins does not involve HGNC. The International Protein Nomenclature Guidelines were written with the involvement of the HGNC.
Nomenclature for regulatory elements: such as promoters, enhancers and transcription-factor binding sites.
Nomenclature for human loci associated with clinical phenotypes and complex traits: The naming of these loci is covered by Online Mendelian Inheritance in Man (OMIM)
Gene naming guideline
HGNC follows a strict set of rules to define gene symbols and gene names. The major key factors are summarized in the table below:

Table 1. Key factors in assigning gene names provided by HGNC (source: Bruford, E.A et al. Nat Genet 52, 754–758 (2020)).
Information for use in data science
Each gene featured within the HGNC database is assigned with a unique identifier or HGNC ID, that is linked to the gene sequence. As a consequence, the HGNC ID will remain constant even if the nomenclature is changed. Although HGNC gene symbols are supposedly constant, these can undergo modifications in exceptional circumstances. For this reasons the HGNC ID is used to unambiguously identify the gene of interest.
Alternate gene symbols (or aliases), locus types (genetic class according to HGNC classification) and chromosomal locations can be accessed by referring to a gene using its identifier.
Implementation in RDF for SPHN
HGNC in RDF is prepared by the SPHN DCC as follows:
download HGNC data from HGNC
translate the data to RDF
The namespace used is: https://www.genenames.org/data/gene-symbol-report/#!/hgnc_id/
A version IRI is provided for each version of HGNC in RDF which indicates the version (or release) of HGNC.
For example, https://biomedit.ch/rdf/sphn-resource/hgnc/20221107 indicates that
the original data was downloaded on 2022-11-07.
For the RDF implementation of the HGNC nomenclature, the following fields have been made available (Table 2):
HGNC ID |
Unique HGNC internal identifier. |
|---|---|
Approved symbol |
Unique symbol approved by HGNC and based on the HGNC nomenclature guidelines. |
Approved name |
Official gene name approved by HGNC in accordance with the HGNC nomenclature guidelines |
Status |
Status of an HGNC record. |
Locus type |
Description of the type of locus associated to the record. |
Locus group |
Group labels that gather similar locus types. |
Alias symbols |
When available, list of additional symbols, based on literature, used to refer to the gene. |
Alias names |
When available, list of additional names, based on literature, used to refer to the gene. |
Accession numbers |
When available, curated list of accession numbers used to link to external databases (e.g., GeneBank). |
Chromosome |
Chromosomal location where the gene lies using cytogenetic coordinates. |
Enzyme ID |
When available, list of identifiers given by the Enzyme Commission to gene products having enzymatic properties. |
NCBI Gene ID |
When available, curated identifier enabling the link with NCBI gene entries. |
Ensembl gene ID |
When available, curated identifier enabling the link with Ensembl gene entries. |
Mouse genome database ID |
When available, curated list of identifiers enabling the link with the Mouse Genomes Informatics (MGI) database gene entries. |
Pubmed ID |
When available, list of identifiers pointing at published articles relevant to the gene entry and featured on Pubmed. |
RefSeq ID |
When available, curated identifier enabling the link with NCBI’s Reference Sequence (RefSeq) collection. Only one selected RefSeq is displayed per gene report. |
Gene group ID |
List of internal HGNC identifiers used to refer to a specific gene group. |
Gene group name |
Gene groups names associated to a gene group identifier. |
CCDC ID |
When available, list of identifiers enabling the link with the Consensus CDS (CCDS) project database. This allows to couple the gene entry with high quality annotated coding regions. |
Vega ID |
When available, curated identifier enabling the link with VEGA entries. |
Table 2. List of HGNC fields available in the current SPHN RDF implementation.
In HGNC, a gene is represented with the following structure:
hgnc:20001 a owl:Class ;
rdfs:label "PCSK9" ;
RO:0002162 NCBITaxon:9606 ;
RO:0002350 hgnc.genegroup:973 ;
RO:0002525 "1p32.3" ;
dc:description "proprotein convertase subtilisin/kexin type 9" ;
oboInOwl:Synonym "FH3",
"NARC-1" ;
oboInOwl:hasDbXref ensembl:ENSG00000169174,
MGI:2140260,
NCBIGene:255738,
genbank:AX207686,
refseq:NM_174936,
vega:OTTHUMG00000008136,
ccds:CCDS603 ;
rdfs:comment "Approved" .
Usage rights
HGNC is jointly published by the US National Human Genome Research Institute (NHGRI) and Wellcome (UK).
The copyright follows the instructions provided by the EMBL-EBI, Wellcome Genome Campus, Hinxton, CB10 1SD. For more details on the usage rights, please consult the EMBL-EBI terms of use (https://www.ebi.ac.uk/about/terms-of-use).
ICD-10-GM
Coding of medical care diagnoses in Switzerland uses the International Statistical Classification Of Diseases And Related Health Problems, 10th revision, German Modification (ICD-10-GM). The German Modification is based on the WHO version and is produced (and updated annually) by the German Federal Institute of Drugs and Medical Devices (BfArM). It is published on the BfArM Website. In Switzerland, a new version is adopted every two years. The Swiss Federal Office of Statistics (BFS) publishes French (called “CIM-10-GM”) and Italian language versions of the ICD-10-GM. See Comparison of versions between SPHN, BFS and BfArM for more information.
The hierarchy of ICD-10-GM is divided into five levels of classification, as shown in Figure 8, with the top level being the most generic and the lowest level the most specific:
Kapitel(chapter) represents the highest level. There are 22 chapters in ICD-10-GM 2021,Gruppe(block) represent the second level of information corresponding to a code range associated with a chapter,Kategorie, Dreistelle(category) represents a three-character code which is the central element of ICD-10-GM,Subkategorie, Viersteller(subcategory) represents a four-character code separated by a decimal point,Subkategorie, Fünfsteller(subcategory) represents a five-character code and is the most specific level of information possible.
Figure 8. Example of ICD-10-GM hierarchy. The chapter code 17 represents the most generic level
of information while the code Q89.00 is the most specific level of information corresponding
to a fifth level code subcategory.
The different levels enable building up the hierarchy of the classification:
the lower the level the more specific the information.
Note: A three-character code can have a four- or five-character code as children.
Comparison of versions between SPHN, BFS and BfArM
The following table provides a side-by-side comparison of versions of ICD-10-GM between SPHN, Swiss Federal Office of Statistics (BFS), and German Federal Institute of Drugs and Medical Devices (BfArM).
SPHN version of ICD-10-GM |
BFS version of ICD-10-GM |
BfArM version of ICD-10-GM |
|---|---|---|
2014 |
2014 |
2012 |
2015 |
2015 |
2014 |
2016 |
2016 |
2014 |
2017 |
2017 |
2016 |
2018 |
2018 |
2016 |
2019 |
2019 |
2018 |
2020 |
2020 |
2018 |
2021 |
2021 |
2021 |
2022 |
2022 |
2022 |
2023 |
2023 |
2022 |
2024 |
2024 |
2022 |
The SPHN release of ICD-10-GM will always be the same version as the one from BFS. For example, from the above table we see that SPHN ICD-10-GM 2023 release is the same as the one from BFS.
But it is entirely possible that the latest version from BFS will not correspond to the latest version from BfArM. For example, from the above table we can see that BFS ICD-10-GM 2023 release actually corresponds to BfArM ICD-10-GM 2022.
Note
This has some consequences, especially for users who are using SPHN ICD-10-GM 2023 but might be relying on the BfArM ICD-10-GM Browser (https://www.dimdi.de/static/de/klassifikationen/icd/icd-10-gm/kode-suche/htmlgm2023/).
For any questions or concerns, please contact the SPHN Data Coordination Center (DCC) at dcc@sib.swiss.
Information for use in data science
ICD-10 is primarily designed to support statistical analysis. ICD-10-GM as well as the WHO version is organized as a mono-hierarchy and provides a classification of diagnoses based on 22 different broad classes of diseases (chapters) numbered using Roman numerals. Each chapter is subdivided into several more specific categories. For example:
chapter I contains certain infectious and parasitic diseases,
chapter X contains diseases of the respiratory system and
chapter XI contains diseases of the digestive system.
Each disease is allocated to only one category. That is, an infectious disease that affects the respiratory and digestive systems is allocated to only one of these 3 chapters. For example, respiratory tuberculosis (code A16) can be found in chapter I, while influenza (codes J09-J18) can be found in chapter X. ICD-10 is therefore useful for any analysis in which it is important that each disease is counted only once. It is less useful for any analyses where there is a need to identify all concepts with a defined meaning. Based on the example above, a query to identify all respiratory infectious diseases would need to include both chapters I and X.
Implementation in RDF for SPHN
The ICD-10-GM is made available in three different CSV files containing altogether the five levels
of information (icd10gm2021syst_kapitel.txt, icd10gm2021syst_gruppen.txt and icd10gm2021syst_kodes.txt).
These files have been parsed (via a Python script) to reconstruct and translate the ICD-10 hierarchy into an RDF representation.
We translate the German (ICD-10-GM) and the French (CIM-10-GM) language versions into RDF.
The namespace used is: <https://biomedit.ch/rdf/sphn-resource/icd-10-gm/>
A version IRI is provided for each version of ICD-10-GM in RDF (e.g. https://biomedit.ch/rdf/sphn-resource/icd-10-gm/2021/1/),
which is composed of the namespace followed by the year of release of ICD-10-GM and the version of the RDF generated for that release.
In the ICD-10-GM RDF, three types of information are stored:
The ICD-10-GM code itself, which is defined as a class (
rdfs:class),language specific names (label) of the ICD-10-GM code are represented by the property
rdfs:label,The hierarchy of the codes, represented by the property
rdfs:subClassOf.
ICD-10-GM versions from 2014 to 2022 have been translated into RDF.
Usage rights
The ICD-10-GM RDF file has been produced using the ICD-10-GM machine-readable version of the BFS.
The copyright is: BFS, Neuchâtel 2021 Wiedergabe unter Angabe der Quelle für nichtkommerzielle Nutzung gestattet (translated to: “Reproduction permitted for non-commercial use provided the source is acknowledged”).
LOINC
Logical Observation Identifier Names and Codes (LOINC, https://loinc.org/) is a common language for identifying health measurements, observations, and documents. LOINC is released twice a year by the Regenstrief Institute, a non-profit clinical research organization. LOINC is publicly available at no cost and provides universal codes and descriptions allowing humans and machines to uniquely identify laboratory tests (e.g. Fasting glucose in Capillary blood) and clinical observations (e.g. Heart rate). With the LOINC Document Ontology, LOINC provides consistent semantics for documents exchanged between systems. The LOINC Document Ontology is not currently used in SPHN.
A LOINC code is made up of the elements shown in Figure 9:
Component: the substance or entity being measured or observed,Property: the characteristic or attribute of the analyte,Time: the timing aspect of the measurement or observation,System: the specimen or thing upon which the observation was made,Scale: how the observation value is quantified or expressed: quantitative, ordinal, nominal,Method: a high-level classification of how the observation was made (optional).
Figure 9. Example of the LOINC code 2340-8. The six different types of information provided by the code are represented. When put in a certain order, these information make up the LOINC Fully-Specified Name (FSN).
LOINC is used as a recommended standard for Lab Tests (https://www.biomedit.ch/rdf/sphn-schema/sphn#LabTest) and for meaning binding section to SPHN concepts.
Information for use in data science
Multicenter research projects using LOINC coded data from different sources can identify similarities and differences in those data. The meaning of the data is readable from the information behind the LOINC code. Using SPARQL queries (see Example of a SPARQL query) a researcher can access this information and gains an important prerequisite for evaluating the comparability of the data.
Example 1: The following LOINC coded data are provided by two different data providers:
Data provider A: 224 records with LOINC code
718-7 Hemoglobin [Mass/volume] in Blood;Data provider B: 310 records with LOINC code
55782-7 Hemoglobin [Mass/volume] in Blood by Oximetry.
Both sets of data come from the measurement of Hemoglobin in Blood in mass concentration.
Five of the six elements are directly comparable.
The difference between the two sets of data is that data provider A
did not specify the measurement method in the code, while data provider B
explicitly specified the method as (Oximetry).
Whether the results of both laboratory tests can be considered and compared
to answer a research question depends on the granularity of information needed.
Example 2: The following LOINC coded data are provided by two different data providers:
Data provider A: 95 records with LOINC code
1556-0 Fasting glucose [Mass/volume] in Capillary blood;Data provider B: 110 records with LOINC code
51596-5 Glucose [Moles/volume] in Capillary blood.
Assuming that there is no further data on eating behavior prior to glucose measurement available the results of the two laboratory tests would not be comparable. The data from data provider A refer to glucose tests taken from patients not allowed to eat anything before the test, while the test information from data provider B does not reveal the pre-conditions (if any). Additional data points outside the LOINC code from data provider B would be needed to assess comparability.
LOINC does not provide codes for machines or test kits and this may also have an impact on the comparability of laboratory test results.
Implementation in RDF for SPHN
The full list of LOINC terms is available as a table at https://loinc.org/downloads/loinc-table/. The LOINC Table Core has been used to generate the RDF representation via a Python script.
The namespace used in the RDF to refer to the LOINC terms is: <https://loinc.org/rdf/>. Each LOINC term is therefore already dereferenceable since the namespace connects to the official webpage of the LOINC classification. Data providers must use this namespace when referring to LOINC terms.
A version IRI is provided for each version of LOINC in RDF (e.g., https://biomedit.ch/rdf/sphn-resource/loinc/2.69/1/),
which is composed of the namespace followed by the version of release of LOINC and
the version of the RDF generated for that release.
However, since LOINC is not providing a generic code for grouping all LOINC terms together,
a LOINC class has been created using the namespace <https://biomedit.ch/rdf/sphn-resource/loinc/>.
A version IRI is provided for each version of LOINC in RDF (e.g., https://biomedit.ch/rdf/sphn-resource/loinc/2021/1/),
which is composed of the namespace followed by the year of release of LOINC and the version of the RDF generated for that release.
Note also that LOINC provides metadata with 6-axis of contextual information for each term. These contextual information have been incorporated in RDF leading to the creation of six properties (e.g., ‘hasComponent’, ‘hasMethodType’) using the SPHN LOINC namespace, <https://biomedit.ch/rdf/sphn-resource/loinc/>. This means that the data user must carefully use the namespaces:
https://loinc.org/rdf/when querying LOINC terms;https://biomedit.ch/rdf/sphn-resource/loinc/when referring to the 6-axis properties.
This release contains a flat list of the LOINC terms in RDF. No hierarchy has been established to cluster the terms under different classes.
Example of a SPARQL query
Many LOINC codes representing Glucose in blood are available.
Using the LOINC Part (https://loinc.org/get-started/loinc-term-basics/),
it is possible to query all LOINC codes which have the Component Glucose and the System Blood called Bld in LOINC:
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX sphn-loinc: <https://biomedit.ch/rdf/sphn-resource/loinc/>
select ?iri ?name_of_the_code
where {
?iri rdfs:label ?name_of_the_code .
?iri sphn-loinc:hasComponent "Glucose" .
?iri sphn-loinc:hasSystem "Bld" .
} limit 100
?iri |
?name_of_the_code |
|---|---|
loinc:15074-8 |
“Glucose [Moles/volume] in Blood” |
loinc:2339-0 |
“Glucose [Mass/volume] in Blood” |
loinc:2340-8 |
“Glucose [Mass/volume] in Blood by Automated test strip” |
loinc:2341-6 |
“Glucose [Mass/volume] in Blood by Test strip manual” |
loinc:5914-7 |
“Glucose [Presence] in Blood by Test strip” |
loinc:72516-8 |
“Glucose [Moles/volume] in Blood by Automated test strip” |
Usage rights
The copyright follows the instructions provided by LOINC (http://loinc.org). LOINC is copyright © 1995-2022, Regenstrief Institute, Inc. and the Logical Observation Identifiers Names and Codes (LOINC) Committee and is available at no cost under the license at http://loinc.org/license. LOINC® is a registered United States trademark of Regenstrief Institute, Inc.
OBI
The Ontology for Biomedical Investigations (OBI) is an ontology that provides terms with precisely defined meanings to describe all aspects of how investigations in the biological and medical domains are conducted. OBI re-uses ontologies that provide a representation of biomedical knowledge from the Open Biological and Biomedical Ontologies (OBO) project and adds the ability to describe how this knowledge was derived.
Information for use in data science
The Ontology for Biomedical Investigations (OBI) is an ontology tha helps you communicate clearly about scientific investigations. Thus, OBI can be used to represent metadata about entities like study, sample, experiment and assay. OBI helps in standardizing terminologies and concepts used to describe experiments, data, and protocols in the field of biomedicine and thus facilitating data integration and knowledge sharing among researchers and institutions.
In the context of SPHN, OBI is used to represent metadata about Assay, Sequencing and Data Processing.
Implementation in RDF for SPHN
OBI is made available as-is by the SPHN DCC.
The namespace used is: http://purl.obolibrary.org/obo/OBI_
A versionIRI` is provided for each version of OBI in RDF which indicates the version (or release) of OBI.
For example, http://purl.obolibrary.org/obo/obi/2023-09-20/obi.owl indicates that the ontology is from a
2023-09-20 release of OBI.
In OBI, a concept is defined with the following structure:
OBI:0001902 a owl:Class ;
rdfs:label "sample preparation for sequencing assay"@en ;
IAO:0000111 "sample preparation for sequencing assay"@en ;
IAO:0000114 IAO:0000120 ;
IAO:0000115 "A sample preparation for assay that preparation of nucleic acids for a sequencing assay" ;
IAO:0000117 "PERSON: Chris Stoeckert, Jie Zheng" ;
IAO:0000119 "NIAID GSCID-BRC metadata working group" ;
OBI:0001886 "Nucleic Acid Preparation Method" ;
dc:source "NIAID GSCID-BRC" ;
rdfs:subClassOf OBI:0000073 ;
owl:equivalentClass [ a owl:Class ;
owl:intersectionOf ( OBI:0000073 [ a owl:Restriction ;
owl:onProperty OBI:0000293 ;
owl:someValuesFrom [ a owl:Class ;
owl:intersectionOf ( OBI:0100051 [ a owl:Restriction ;
owl:onProperty OBI:0000643 ;
owl:someValuesFrom CHEBI:33696 ] ) ] ] [ a owl:Restriction ;
owl:onProperty OBI:0000299 ;
owl:someValuesFrom [ a owl:Class ;
owl:intersectionOf ( OBI:0100051 [ a owl:Restriction ;
owl:onProperty OBI:0000295 ;
owl:someValuesFrom OBI:0600047 ] ) ] ] ) ] .
Usage rights
OBI is accessible in the public domain under the CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/).
ORDO
The Orphanet Rare Disease Ontology (ORDO) is a structured vocabulary for rare diseases derived from the Orphanet database, capturing relationships between diseases, genes and other relevant features. ORDO provides integrated, re-usable data for computational analysis of rare diseases.
Information for use in data science
ORDO is derived from the Orhpanet database and integrates classification of rare diseases, gene-disease relationships, epidemiological data, and connections to other terminologies (like MeSH, UMLS, MedDRA), databases (like OMIM, UniProtKB, HGNC, Ensembl, Reactome, IUPHAR, GeneAtlas) and classifications (like ICD-10).
In the context of SPHN, ORDO is used to represent information about Disorder and Diagnosis.
Implementation in RDF for SPHN
ORDO is made available as-is by the SPHN DCC.
The namespace used is: http://www.orpha.net/ORDO/Orphanet_
A versionIRI is provided for each version of OBI in RDF which indicates the version (or release) of ORDO.
For example, https://www.orphadata.com/data/ontologies/ordo/last_version/ORDO_en_4.4.owl indicates that the ontology is from the 4.4 release of ORDO.
In ORDO, a concept is defined with the following structure:
Orphanet:1251 a owl:Class ;
rdfs:label "Blepharofacioskeletal syndrome" ;
efo:alternative_term "Richieri Costa-Guion Almeida-Rodini syndrome"@en ;
efo:reason_for_obsolescence "This entity has been excluded from the Orphanet nomenclature of rare diseases and moved to Schilbach-Rott syndrome" ;
Orphanet:C055 "https://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=en&Expert=1251" ;
rdfs:subClassOf [ a owl:Restriction ;
owl:onProperty Orphanet:C056 ;
owl:someValuesFrom Orphanet:2353 ],
Orphanet:C044 ;
skos:notation "ORPHA:1251" .
Usage rights
ORDO is accessible in the public domain under the CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/).
SNOMED CT
Systematized Nomenclature of Medicine Clinical Terms – SNOMED CT (https://www.snomed.org/) is a global standard for health terms and a common language designed for use in Electronic Health Records (EHRs). The international edition of SNOMED CT is released monthly by the International Health Terminology Standards Development Organization (IHTSDO). Switzerland holds a license of SNOMED CT and Swiss organizations can use SNOMED CT free of charge when registering with the Swiss National Release Center (NRC) eHealth Suisse.
Note
In addition to the international edition of SNOMED CT, there is a Swiss extension prepared by the eHealth Suisse. The extension consists of new concepts that are specific to Switzerland. From 2024 onwards, the SPHN DCC will provide the international edition of SNOMED CT combined with the Swiss extension from eHealth Suisse.
SNOMED CT is published in RF2 format and can be explored on the web through https://browser.ihtsdotools.org. IHTSDO provides regular training courses as well as a library with training videos on all topics, which are available to anyone after free registration on SNOMED CTs e-learning platform (https://elearning.ihtsdotools.org/).
Note
The Swiss extension of SNOMED CT is published in RF2 format by the eHealth Suisse and can be explored on the web through the IHTSDO Browser.
SNOMED CT provides unique identifiers for concepts (clinical ideas), which are defined though is-a relationships and attribute relationships to other concepts. It is providing different descriptions with their own identifiers such as synonyms, and it is organized in a polyhierarchical manner which means that a concept can have multiple parents.
Information for use in data science
SNOMED CT can be used for analytics of structured and unstructured data, e.g. querying clinical data using the machine processable concept definitions defined in SNOMED CT, or using SNOMED CT for analyzing free text with Natural Language Processing (NLP) tools. In this fact sheet we focus on analytics of structured data. SNOMED CT features described above allow queries using the SNOMED CT hierarchies (e.g. body structure, substance, clinical finding) as well as so called attribute relationships (e.g. causative agent, pathological process). Further, SPHN defines value sets based on SNOMED CT which can help defining query criteria.
Implementation of SNOMED CT in RDF
SNOMED CT in RDF has been generated thanks to the Snomed OWL Toolkit.
The OWL produced by the Snomed OWL Toolkit outputs the data to owl functional syntax. This format is readable by some ontology editors but not all databases. Therefore DCC provides conversion to other formats such as Turtle or ntriples.
The namespace provided in the RDF to refer to SNOMED CT terms is: <https://snomed.info/id/>.
The ontology IRI defined by SNOMED is: <http://snomed.info/sct/900000000000207008> A version IRI is provided for each version in the form of: <http://snomed.info/sct/900000000000207008/version/20221231> where the last part is composed of the date in the form of yyyyMMdd of the release.
In SNOMED CT several information is stored:
The OWL conversion implements every concept as an
owl:classand properties asowl:ObjectProperty.Several
rdfs:subClassOfare constructing the hierarchy of the classes andrdfs:subPropertyOfin the properties.Names are annotated using
rdfs:labelas well asskos:altLabelandskos:prefLabel. The annotations are localized typically with the language tag@en. In some cases there exist alternative labels in@en-gbor@en-us.owl:equivalentClassorrdfs:subClassOfis used when there is an overlap with other classes.
Example of a SPARQL query
The SPHN dataset defines that SNOMED CT is used as a standard to express the allergen or
the substance that triggered an allergy episode in a patient.
It is further defined that the SNOMED CT concept identifying the substance must be a sub concept of
SNOMED CT concept 105590001 |Substance (substance)|.
Connecting biomedical data with SNOMED CT allows for queries using the SNOMED CT hierarchies.
For our example illustrating one of these hierarchy paths we imagine that for some patients
an allergy episode for the substance Peanut is recorded,
and a researcher is interested to query for all patients that experienced an allergy episode
due to consumption of legume (synonym of Pulse vegetable).
The code block below presents an example data with information about four patients,
where three of them have an allergy: two are annotated to be allergic to Pulse vegetable and one to Peanut specifically:
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX snomed: <http://snomed.info/id/>
PREFIX sphn: <https://biomedit.ch/rdf/sphn-ontology/sphn/>
PREFIX resource: <http://biomedit.ch/rdf/sphn-resource/>
resource:patient123 a sphn:SubjectPseudoIdentifier ;
sphn:hasIdentifier "patient123" .
resource:patient234 a sphn:SubjectPseudoIdentifier ;
sphn:hasIdentifier "patient234" .
resource:patient345 a sphn:SubjectPseudoIdentifier ;
sphn:hasIdentifier "patient345" .
resource:patient456 a sphn:SubjectPseudoIdentifier ;
sphn:hasIdentifier "patient456" .
resource:allergyEpisode123 a sphn:AllergyEpisode ;
sphn:hasAllergen resource:allergen1 ;
sphn:hasSubjectPseudoIdentifier resource:patient123 .
resource:allergyEpisode234 a sphn:AllergyEpisode ;
sphn:hasAllergen resource:allergen2 ;
sphn:hasSubjectPseudoIdentifier resource:patient234 .
resource:allergyEpisode456 a sphn:AllergyEpisode ;
sphn:hasAllergen resource:allergen1 ;
sphn:hasSubjectPseudoIdentifier resource:patient456 .
resource:allergen1 a sphn:Allergen ;
sphn:hasCode resource:allergen1-code .
# Pulse vegetable
resource:allergen1-code a snomed:227313005 .
resource:allergen2 a sphn:Allergen ;
sphn:hasCode resource:allergen2-code .
# Peanuts
resource:allergen2-code a snomed:762952008 .
snomed:762952008 rdfs:subClassOf snomed:227313005.
A simple query enables to retrieve patients allergic to a substance in the family of Pulse Vegetable:
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX snomed: <http://snomed.info/id/>
PREFIX sphn: <https://biomedit.ch/rdf/sphn-ontology/sphn/>
PREFIX resource: <http://biomedit.ch/rdf/resource/>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
select DISTINCT ?patient
where {
# Allergy episodes and their substance codes:
?allergy rdf:type sphn:AllergyEpisode .
?allergy sphn:hasAllergen ?allergen .
?allergen sphn:hasCode ?code .
# Patients linked to these episodes:
?allergy sphn:hasSubjectPseudoIdentifier ?patient .
# Substance code should be a pulse vegetable (snomed:227313005) or any descendant:
?code rdf:type ?pulse_veg_and_descendants .
?pulse_veg_and_descendants rdfs:subClassOf* snomed:227313005 .
}
Therefore, the query returns the three patients even if one was specifically allergic to Peanuts:
?patient |
|---|
patient123 |
patient234 |
patient456 |
This is due to the hierarchical structure of ontology and the reasoning possibilities offered by semantic graph technologies. For the query above the SNOMED CT substance hierarchy is important, namely the concept Pulse vegetable and the concept Peanut which is a direct descendent of Pulse vegetable in SNOMED CT.
105590001 |Substance (substance)|. . .227313005 |Pulse vegetable (substance)|762952008 |Peanut (substance)|
Usage rights
The copyright follows the instructions provided by SNOMED CT (https://www.snomed.org/), SNOMED CT is copyright © SNOMED International 2021 v3.15.1., SNOMED CT international. In order to use the file please register with eHealth Suisse for an affiliate license for SNOMED CT (free of charge) https://mlds.ihtsdotools.org/#/landing/CH?lang=en.
SO
The Sequence Ontology (SO, http://www.sequenceontology.org/) is a structured controlled vocabulary that aims at facilitating the exchange, analysis and organization of genomic data by humans and machines alike. For its development, the ontology rallied contributions from different communities such as the Generic Model Organism Database (GMOD), the Sanger Institute and the European Bioinformatics Institute (EBI) group as well as from widely used model organism databases (e.g., WormBase, FlyBase, Mouse Genome Informatics group).
The SO is designed as a tool that unifies the way in which sequence annotations are described. To achieve this goal, it relies on several terms that describe parts of the sequence annotations, as well as the relationships between these. The use of a common controlled vocabulary during the annotation process enables the comparisons between annotations from different project and ease its downstream analysis.
Information for use in data science
Each term provided by the Sequence Ontology (SO) is linked to an accession (or concept identifier), a human-readable definition and its source as seen in Figure 10A. These not only describe common genomic annotations (e.g., exon, intron, binding_site), but also experimental features (e.g., microarray_oligo, smFISH_probe) thus allowing to connect the sequence and its biology to the results of an experiment.
In the SO, the terms are linked by a set of relationships that allow to perform logical inferences about the annotated data. In addition, the definition of clear relationships between terms introduce restrictions in the way the annotation is performed.
For example, in SO an ‘Exon’ is part of a ‘Transcript’ whereas an ‘Intron’ is part of a ‘Primary transcript’ which is a ‘Transcript’, as seen in Figure 10B. It is however incorrect to state that an ‘Intron’ is part of a ‘Transcript’. These clear relationships ultimately help to maintain consistency across different projects annotations.
Figure 10. Terms and their relationships in SO. A) Each term is identified by its accession number, a description, synonyms and eventual cross references, as well as all direct parents and children terms linked to it. B) The relationship between terms (arrows) are labelled as follows: (i) indicates an “is_a” relationship. (P) indicates a “part_of” relationship. (d) indicates a ‘derived_from” relationship (source: Eilbeck, K. et al. Genome Biol 6, R44 (2005)).
The ontology relies on three types of relationships, namely:
is_a: allows to represent hierarchies (i.e., mRNA is_a Processed transcript)
derived_from: implies a precise relationship between the terms (i.e., polypeptide derive_from mRNA)
part_of: allows to represent part-whole relationships between terms (i.e., exon is a part_of transcript)
Implementation in RDF for SPHN
The SO is made available as-is by the DCC.
The namespace used is: <http://purl.obolibrary.org/obo/SO_>
A version IRI is provided for each version of SO in RDF which indicates the version (or release) of SO.
For example, http://purl.obolibrary.org/obo/so/2021-11-22/so.owl indicates that the ontology is from a 2021-11-22 release of SO.
In SO, a concept is defined with the following structure:
SO:0000704 a owl:Class ;
rdfs:label "gene"^^xsd:string ;
IAO:0000115 "A region (or regions) that includes all of the sequence elements necessary to encode a functional transcript. A gene may include regulatory regions, transcribed regions and/or other functional sequence regions."^^xsd:string ;
oboInOwl:hasDbXref "http://en.wikipedia.org/wiki/Gene"^^xsd:string ;
oboInOwl:hasExactSynonym "INSDC_feature:gene"^^xsd:string ;
oboInOwl:hasOBONamespace "sequence"^^xsd:string ;
oboInOwl:id "SO:0000704"^^xsd:string ;
oboInOwl:inSubset so:SOFA ;
rdfs:comment "This term is mapped to MGED. Do not obsolete without consulting MGED ontology. A gene may be considered as a unit of inheritance."^^xsd:string ;
rdfs:subClassOf [ a owl:Restriction ;
owl:onProperty so:member_of ;
owl:someValuesFrom SO:0005855 ],
SO:0001411 .
Usage rights
SO is maintained by the Eilbeck Lab, Department of Biomedical Informatics, University of Utah, Salt Lake City.
SO data and data products are licensed under the Creative Commons Attribution 4.0 Unported License (http://www.sequenceontology.org/?page_id=345)
UCUM
The Unified Code for Units of Measure (UCUM, https://ucum.org/trac) is a compositional code system intended to
include all units of measures being contemporarily used in international science, engineering, and business.
UCUM units can be composed by using the UCUM expression syntax.
UCUM, similarly to LOINC, is released and maintained by the Regenstrief Institute.
It is adopted by standard organization such as DICOM (https://www.dicomstandard.org/)
or HL7 (https://www.hl7.org/) and is strongly encouraged by LOINC (https://loinc.org/).
UCUM is used as one recommended standard for the SPHN concept Unit (https://www.biomedit.ch/rdf/sphn-schema/sphn#Unit).
Technical specification
Next to standard units such as L=liters, g=grams, UCUM allows for:
Prefixes e.g.
kfor ‘kilo’ orufor ‘micro’Annotations in
{}e.g.mU/g {Hgb}for ‘miliUnits per gram Hemoglobin’Scaling factors such as
10*Xe.g.10*3/ulfor ‘thousand per microliter’Lexical elements in
[]e.g.mm[Hg]for ‘millimeters of mercury’Operators e.g.
.for ‘multiplications’ and/for ‘division’
Conventions in use in SPHN
The UCUM clearly defines each available unit with concise semantics, but also allows for the use of free-text annotations that can be added to defined variables or used instead of a unit.
Numerical modifiers
The UCUM is perfectly capable of interpreting numeric modifiers directly associated with a unit. For example, the unit (10*6.mol)/kg or dimensionless unit 10*6 are both resolved without issues by an UCUM interpreter, but present some obvious issues when it comes to comparing values with the same dimension but different numerical modifier.
For this reason, all new units added to the SPHN UCUM RDF release (since UCUM-2023-1) are devoided of numerical modifiers and replaced by their “core” unit. For instance, in the case of the example (10*6.mol)/kg becomes simply mol/kg.
Note: Existing units with numerical modifiers (pre UCUM-2023-1) are not deprecated.
There are a few exceptions where new units with numerical modifiers are accepted. This occurs when the numerical modifier is an integral part of the core unit, and the value recalculation would introduce an error. This is the case for units like g/(100.mL) where correcting the values (e.g., dividing all values by 100) would introduce errors due to the approximations made during the measurement itself.
Usage of UCUM annotations
The UCUM clearly defines each available unit with concise semantics, but also allows for the use of free-text annotations that can be added to defined variables or used instead of a unit.
Although this practice is discouraged, it is widely used in several scientific fields, such as clinical research. To address this issue, SPHN has developed a series of recommendations for the use of UCUM annotations when deemed necessary:
Use of the English language
Avoid abbreviations
Use of the singular form
Use of lower-case spelling
Example: The French term ‘Spécimen’, often used as part of units such as ‘µg/Spécimen’ would become ug/{specimen}
Dimensionless units
Dimensionless units are measurements devoid of physical dimensions, signifying a lack of association with specific physical quantities. This holds true for ratios, counts, or percentages.
While UCUM enables the representation of percentages using the symbol % or common ratios as fractions of the same core unit, such as kg/kg or mol/mol it does not provide a straightforward solution for expressing counts.
For this reason, SPHN recommends the use of {#} to express any form of counting.
Implementation in RDF for SPHN
For the SPHN RDF file, we did not implement full support for the syntax. We rely on a large library of pre-built valid UCUM codes from the National Library of Medicine, National Institutes of Health, U.S. Department of Health and Human Services with content contributions from Intermountain Healthcare and the Regenstrief Institute (download list, and additional information, Version 1.5, Released 06/2020.).
In addition, a couple of units from the SPHN dataset (e.g. cGy, MBq, mCi), not included in this library, has been added.
The namespace used in the RDF to refer to the UCUM terms is: <http://biomedit.ch/sphn-resource/ucum/>.
Since UCUM does not have a release cycle, the versioning relies on the year when the RDF is generated by the DCC. The version IRI is provided for each version of UCUM in RDF (e.g., https://biomedit.ch/rdf/sphn-resource/ucum/2021/1/), which is composed of the namespace followed by the year the RDF is being generated and the version of the RDF generated for that year.
In this RDF, a root class is generated (https://biomedit.ch/rdf/sphn-resource/ucum/UCUM).
The UCUM codes are provided as rdfs:Class, with a rdfs:label indicating the UCUM code,
a rdfs:comment indicating the human-readable meaning of the code and the codes are sublasses to the ucum:UCUM
root class (see example below of the unit kilogram):
ucum:kg a rdfs:Class ;
rdfs:label "kg" ;
rdfs:comment "kilogram" ;
rdfs:subClassOf ucum:UCUM .
There hasn’t been any hierarchy established to cluster the terms under different classes.
If you need an UCUM code not present in the list, please submit a request to dcc@sib.swiss, we could include it in the next SPHN UCUM RDF release.
Note
In the context of SPHN, UCUM codes are being defined as rdfs:Class only since 2024. Before, they were defined as owl:NamedIndividuals.
Usage rights
The copyright follows the instructions provided by Regenstrief Institute. UCUM is Copyright © 1999-2013 Regenstrief Institute, Inc. and The UCUM Organization, Indianapolis, IN. All rights reserved. See TermsOfUse.
Other terminologies
There exist other external resources (e.g., GUDID, ICD-O-3) recommended in the SPHN Dataset for coding values in SPHN besides the ones introduced in the previous sections. These resources are not necessarily provided in the RDF format by the DCC as a single .ttl or .owl file for use.
For these resources, the DCC recommends to provide the codes in the RDF data files with the Code concept with the following attributes:
the identifier
the name
the coding system and version (following conventions defined here)
For example, if an GUDID code 04015630935406 is provided, the following data instance must be generated by the data provider:
resource:Code-GUDID-04015630935406 a sphn:Code ;
sphn:hasIdentifier "04015630935406"^^xsd:string ;
sphn:hasName "cobas 6800 System"^^xsd:string ;
sphn:hasCodingSystemAndVersion "GUDID"^^xsd:string .
Find below a (non-exhaustive) list of such external resources with an introduction and possibly information about their use in data science.
GUDID
Introduction to the classification
The Global Unique Device Identification Database (GUDID) contains key device identification information submitted to the FDA about medical devices that have Unique Device Identifiers (UDI). The FDA is establishing the unique device identification system to adequately identify devices sold in the U.S. - from manufacturing through distribution to patient use.
Source: https://accessgudid.nlm.nih.gov/
GUDID is used as one of the recommended standard for the SPHN concept:
Lab Analyzer (https://www.biomedit.ch/rdf/sphn-schema/sphn#LabAnalyzer).
Information for use in data science
With hospitals and data providers becoming more and more interconnected, e.g., in national or international multi-center projects, data interoperability is of growing importance. The use of LOINC-codes covers clinical observations and laboratory tests, making clinical data collected across different hospitals or laboratories more comparable. LOINCs, however, are often missing information on the method employed to generate a laboratory result, providing no or rather high-level information only. The type of method, analyzer device, or reagent (specified for example by GMDN or EMDN) or even the exact model of a medical device(s) used is of major interest for clinical staff and researchers. When specifying data used to generate reference values for laboratory tests, for example, it is crucial to ensure compatibility of data sources, as therapeutic decisions may rely on them. Specification of devices is unfortunately often still inconsistent between hospitals and laboratories or languages. The use of unique device identifiers (UDIs), e.g., from the Global Unique Device Identifier Database (GUDID) or the upcoming EUDAMED database will be highly beneficial to assign a medical device to a laboratory test without ambiguities.
Example 1
Modern laboratory analyzers are often modular, for example, the Cobas 8000 from Roche featuring several modules for different applications (clinical chemistry and immunoassays) with different rates of throughput. Specifying “Cobas 8000” as medical device used for data collection is therefore not sufficiently granular. Likewise, a human specialist can easily identify designations like “Cobas 8000 c 502”, “Cobas c‑502”, or “c 502 Cobas 8000” as identical, while an automated data processor cannot. A unique identifier (04015630928354) can overcome this issue and in addition link to associated information like manufacturer, catalog number or a device description.
Example 2
Names of laboratory analyses used internally in clinical data warehouses often lack specificity in terms of reagent, test kit or potential supplementary reagents used or required. The assessment of levels of alanine aminotransferase (ALAT), for example, can be done from different sample sources and according to different standards (e.g. with or without addition of pyridoxal phosphate in the reaction). While the former can be specified by the corresponding LOINC (e.g. 1742-6 “serum or plasma” vs. 76625-3 “blood”), the latter can be assigned by the use of a unique identifier for the test kit (04015630920532 “ALAT with pyridoxal phosphate activation” vs. 04015630913862 “ALAT without pyridoxal phosphate activation”).
The combination of product identifiers and type categorization systems like EMDN or GMDN will help to provide information on medical devices with different degrees of granularity, suitable for various purposes.
ICD-O-3
Introduction to the classification
The International Classification of Diseases for Oncology – 3rd Edition (ICD-O-3) is used for coding the body site (topography) and the histology (morphology) of neoplasms. The information is usually obtained from a pathology report and ICD-O-3 is used in cancer registries around the world. ICD-O-3 is provided by the World Health Organization (WHO) and has been updated twice by the International Association of Cancer Registries (IACR) on behalf of WHO (ICD-O-3 1st revision from 2013 and ICD-O-3 2nd revision from 2019).
ICD-O-3 is a multi-axial classification with codes for topography, morphology, behavior and grading of neoplasm (see Figure 11). The topography hierarchy uses mainly the ICD-10 classification of malignant neoplasm. The morphology non-hierarchical axis provides codes composed of histology and behavior. Histologic grading (differentiation) is a separate code.
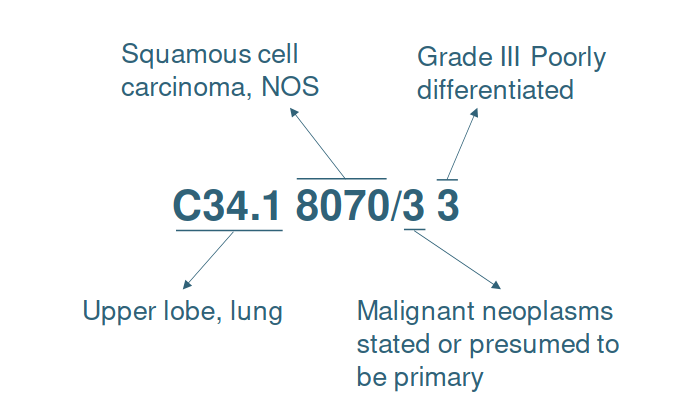
Figure 11. Example of the ICD-O-3 code C34.1 8070/3 3 meaning “Poorly differentiated squamous cell carcinoma, upper lobe of lung”.
ICD-O-3 is used as the recommended standard for the SPHN concept:
ICD-O Diagnosis (https://www.biomedit.ch/rdf/sphn-schema/sphn#ICDODiagnosis).
Versions of ICD-O-3
In Switzerland, ICD-O-3 is used since 2005; the ICD-O-3 1st revision (ICD-O-3.1) was used from 2013 to 2019 and the ICD-O-3 2nd revision (ICD-O-3.2) was used from 2020 onwards.